* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Gene!
Copy-number variation wikipedia , lookup
Oncogenomics wikipedia , lookup
Genomic library wikipedia , lookup
Epigenetics in learning and memory wikipedia , lookup
Public health genomics wikipedia , lookup
Non-coding RNA wikipedia , lookup
Transfer RNA wikipedia , lookup
Gene therapy wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Short interspersed nuclear elements (SINEs) wikipedia , lookup
Genetic engineering wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Pathogenomics wikipedia , lookup
Transposable element wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Biology and consumer behaviour wikipedia , lookup
Gene nomenclature wikipedia , lookup
Gene expression programming wikipedia , lookup
Genomic imprinting wikipedia , lookup
Frameshift mutation wikipedia , lookup
Ridge (biology) wikipedia , lookup
Messenger RNA wikipedia , lookup
Gene desert wikipedia , lookup
Epitranscriptome wikipedia , lookup
Human genome wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Point mutation wikipedia , lookup
Non-coding DNA wikipedia , lookup
History of genetic engineering wikipedia , lookup
Expanded genetic code wikipedia , lookup
Genome (book) wikipedia , lookup
Minimal genome wikipedia , lookup
Primary transcript wikipedia , lookup
Genome editing wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Gene expression profiling wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Microevolution wikipedia , lookup
Designer baby wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Helitron (biology) wikipedia , lookup
Genome evolution wikipedia , lookup
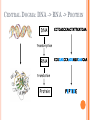
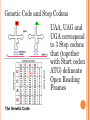
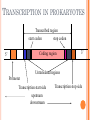
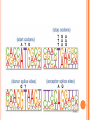
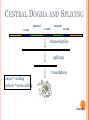
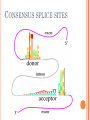
BIOINFORMATICS Lecture 4 Gene Prediction Dr. Aladdin Hamwieh Khalid Al-shamaa Abdulqader Jighly Aleppo University Faculty of technical engineering Department of Biotechnology 2010-2011 GENE PREDICTION: COMPUTATIONAL CHALLENGE Gene: A sequence of nucleotides coding for protein Gene Prediction Problem: Determine the beginning and end positions of genes in a genome GENE PREDICTION: COMPUTATIONAL CHALLENGE aatgcatgcggctatgctaatgcatgcggctatgctaagctgggatccgatgacaatgcatg cggctatgctaatgcatgcggctatgcaagctgggatccgatgactatgctaagctgggatcc gatgacaatgcatgcggctatgctaatgaatggtcttgggatttaccttggaatgctaagctg ggatccgatgacaatgcatgcggctatgctaatgaatggtcttgggatttaccttggaatatg ctaatgcatgcggctatgctaagctgggatccgatgacaatgcatgcggctatgctaatgcat gcggctatgcaagctgggatccgatgactatgctaagctgcggctatgctaatgcatgcggct atgctaagctgggatccgatgacaatgcatgcggctatgctaatgcatgcggctatgcaagc tgggatcctgcggctatgctaatgaatggtcttgggatttaccttggaatgctaagctgggat ccgatgacaatgcatgcggctatgctaatgaatggtcttgggatttaccttggaatatgctaa tgcatgcggctatgctaagctgggaatgcatgcggctatgctaagctgggatccgatgacaa tgcatgcggctatgctaatgcatgcggctatgcaagctgggatccgatgactatgctaagctg cggctatgctaatgcatgcggctatgctaagctcatgcggctatgctaagctgggaatgcatg cggctatgctaagctgggatccgatgacaatgcatgcggctatgctaatgcatgcggctatgc aagctgggatccgatgactatgctaagctgcggctatgctaatgcatgcggctatgctaagct cggctatgctaatgaatggtcttgggatttaccttggaatgctaagctgggatccgatgacaa tgcatgcggctatgctaatgaatggtcttgggatttaccttggaatatgctaatgcatgcggct atgctaagctgggaatgcatgcggctatgctaagctgggatccgatgacaatgcatgcggct atgctaatgcatgcggctatgcaagctgggatccgatgactatgctaagctgcggctatgcta atgcatgcggctatgctaagctcatgcgg GENE PREDICTION: COMPUTATIONAL CHALLENGE aatgcatgcggctatgctaatgcatgcggctatgctaagctgggatccgatgacaatgcatg cggctatgctaatgcatgcggctatgcaagctgggatccgatgactatgctaagctgggatcc gatgacaatgcatgcggctatgctaatgaatggtcttgggatttaccttggaatgctaagctg ggatccgatgacaatgcatgcggctatgctaatgaatggtcttgggatttaccttggaatatg ctaatgcatgcggctatgctaagctgggatccgatgacaatgcatgcggctatgctaatgcat gcggctatgcaagctgggatccgatgactatgctaagctgcggctatgctaatgcatgcggct atgctaagctgggatccgatgacaatgcatgcggctatgctaatgcatgcggctatgcaagc tgggatcctgcggctatgctaatgaatggtcttgggatttaccttggaatgctaagctgggat ccgatgacaatgcatgcggctatgctaatgaatggtcttgggatttaccttggaatatgctaa tgcatgcggctatgctaagctgggaatgcatgcggctatgctaagctgggatccgatgacaa tgcatgcggctatgctaatgcatgcggctatgcaagctgggatccgatgactatgctaagctg cggctatgctaatgcatgcggctatgctaagctcatgcggctatgctaagctgggaatgcatg cggctatgctaagctgggatccgatgacaatgcatgcggctatgctaatgcatgcggctatgc aagctgggatccgatgactatgctaagctgcggctatgctaatgcatgcggctatgctaagct cggctatgctaatgaatggtcttgggatttaccttggaatgctaagctgggatccgatgacaa tgcatgcggctatgctaatgaatggtcttgggatttaccttggaatatgctaatgcatgcggct atgctaagctgggaatgcatgcggctatgctaagctgggatccgatgacaatgcatgcggct atgctaatgcatgcggctatgcaagctgggatccgatgactatgctaagctgcggctatgcta atgcatgcggctatgctaagctcatgcgg GENE PREDICTION: COMPUTATIONAL CHALLENGE aatgcatgcggctatgctaatgcatgcggctatgctaagctgggatccgatgacaatgcatg cggctatgctaatgcatgcggctatgcaagctgggatccgatgactatgctaagctgggatcc gatgacaatgcatgcggctatgctaatgaatggtcttgggatttaccttggaatgctaagctg ggatccgatgacaatgcatgcggctatgctaatgaatggtcttgggatttaccttggaatatg ctaatgcatgcggctatgctaagctgggatccgatgacaatgcatgcggctatgctaatgcat gcggctatgcaagctgggatccgatgactatgctaagctgcggctatgctaatgcatgcggct atgctaagctgggatccgatgacaatgcatgcggctatgctaatgcatgcggctatgcaagc tgggatcctgcggctatgctaatgaatggtcttgggatttaccttggaatgctaagctgggat ccgatgacaatgcatgcggctatgctaatgaatggtcttgggatttaccttggaatatgctaa tgcatgcggctatgctaagctgggaatgcatgcggctatgctaagctgggatccgatgacaa tgcatgcggctatgctaatgcatgcggctatgcaagctgggatccgatgactatgctaagctg cggctatgctaatgcatgcggctatgctaagctcatgcggctatgctaagctgggaatgcatg cggctatgctaagctgggatccgatgacaatgcatgcggctatgctaatgcatgcggctatgc aagctgggatccgatgactatgctaagctgcggctatgctaatgcatgcggctatgctaagct cggctatgctaatgaatggtcttgggatttaccttggaatgctaagctgggatccgatgacaa tgcatgcggctatgctaatgaatggtcttgggatttaccttggaatatgctaatgcatgcggct atgctaagctgggaatgcatgcggctatgctaagctgggatccgatgacaatgcatgcggct atgctaatgcatgcggctatgcaagctgggatccgatgactatgctaagctgcggctatgcta atgcatgcggctatgctaagctcatgcgg Gene! CENTRAL DOGMA: DNA -> RNA -> PROTEIN DNA CCTGAGCCAACTATTGATGAA transcription RNA CCUGAGCCAACUAUUGAUGAA translation Protein PEPTIDE CODONS In 1961 Sydney Brenner and Francis Crick discovered frameshift mutations Systematically deleted nucleotides from DNA Single and double deletions dramatically altered protein product Effects of triple deletions were minor Conclusion: every triplet of nucleotides, each codon, codes for exactly one amino acid in a protein THE SLY FOX In the following string THE SLY FOX AND THE SHY DOG Delete 1, 2, and 3 nucleotifes after the first ‘S’: THE SYF OXA NDT HES HYD OG THE SFO XAN DTH ESH YDO G THE SOX AND THE SHY DOG Which of the above makes the most sense? TRANSLATING NUCLEOTIDES INTO AMINO ACIDS Codon: 3 consecutive nucleotides 43 = 64 possible codons Genetic code is degenerative and redundant Includes start and stop codons An amino acid may be coded by more than one codon Genetic Code and Stop Codons UAA, UAG and UGA correspond to 3 Stop codons that (together with Start codon ATG) delineate Open Reading Frames TERMINOLOGY ORF Exons Portions of the ORF that are transcribed and when combined form the coding sequence (CDS) for the gene Introns A series of DNA codons, including a 5’ initiation codon and a termination codon, that encodes a putative or known gene. Portions of the ORF that are transcribed and are spliced out of the mRNA before translation. Untranslated regions (UTRs) Non-coding regions that are transcribed and flank the ORF (for DNA) and CDS (for mRNA) 5’ end (relative to mRNA) UTRs (leader, regulatory sites) 3’ end UTRs (terminator sites, trailer) TRANSCRIPTION IN PROKARYOTES Transcribed region start codon stop codon Coding region 5’ 3’ Untranslated regions Promoter Transcription start side upstream downstream Transcription stop side GENE STRUCTURE IN EUKARYOTS Transcribed region exons introns start codon stop codon 5’ 3’ donor GT AG acceptor and sites Promoter Transcription stop site Untranslated regions Transcription start site BIOLOGICAL BACKGROUND GENE STRUCTURE 5’ UTR Ex1 In1 Ex2 Stop codon ATG Ex2 In2 Ex3 In3 GT CAP Ex1 Ex2 Ex2 Ex3 Ex4 In4 Ex5 Ex5 Ex5 Poly A AG Ex4 3’ UTR Ex5 DISCOVERY OF SPLIT GENES In 1977, Phillip Sharp and Richard Roberts experimented with mRNA of hexon, a viral protein. Map hexon mRNA in viral genome by hybridization to adenovirus DNA and electron microscopy mRNA-DNA hybrids formed three curious loop structures instead of contiguous duplex segments EXONS AND INTRONS In eukaryotes, the gene is a combination of coding segments (exons) that are interrupted by noncoding segments (introns) This makes computational gene prediction in eukaryotes even MORE DIFFICULT Prokaryotes don’t have introns Genes in prokaryotes are continuous CENTRAL DOGMA AND SPLICING exon1 intron1 exon2 intron2 exon3 transcription splicing exon = coding intron = non-coding translation Batzoglou SPLICING SIGNALS Exons are interspersed with introns and typically flanked by GT and AG CONSENSUS SPLICE SITES TWO APPROACHES TO GENE PREDICTION Statistical: coding segments (exons) have typical sequences on either end and use different subwords than non-coding segments (introns). Similarity-based: many human genes are similar to genes in mice, chicken, or even bacteria. Therefore, already known mouse, chicken, and bacterial genes may help to find human genes. STATISTICAL APPROACH: METAPHOR IN UNKNOWN LANGUAGE Noting the differing frequencies of symbols (e.g. ‘%’, ‘.’, ‘-’) and numerical symbols could you distinguish between a story and the stock report in a foreign newspaper? SIMILARITY-BASED APPROACH: METAPHOR IN DIFFERENT LANGUAGES If you could compare the day’s news in English, side-by-side to the same news in a foreign language, some similarities may become apparent WHICH ISSUES HELP US TO DETECT GENES? 1. 2. 3. 4. 5. 6. 7. ORFs constructions ORFs Long ORFs Codon usage Regulatory motifs Ribosomal Binding Sites CG islands Similarities with other genes OPEN READING FRAMES (ORFS) Detect potential coding regions by looking at ORFs A genome of length n is comprised of (n/3) codons Stop codons break genome into segments between consecutive Stop codons The subsegments of these that start from the Start codon (ATG) are ORFs ORFs in different frames may overlap ATG TGA Genomic Sequence Open reading frame LONG VS.SHORT ORFS Long open reading frames may be a gene At random, we should expect one stop codon every (64/3) ~= 21 codons However, genes are usually much longer than this A basic approach is to scan for ORFs whose length exceeds certain threshold This is naive because some genes (e.g. some neural and immune system genes) are relatively short 9-kb fungal plasmid sequence in looking for ORFs (potential genes). Six possible reading frames, three in each direction. Two large ORFs, 1 and 2, are the most likely candidates as potential genes. The yellow ORFs are too short to be genes TESTING ORFS: CODON USAGE Create a 64-element hash table and count the frequencies of codons in an ORF Amino acids typically have more than one codon, but in nature certain codons are more in use Uneven use of the codons may characterize a real gene This compensate for pitfalls of the ORF length test CODON USAGE IN HUMAN GENOME CODON USAGE IN MOUSE GENOME AA codon Ser Ser Ser Ser Ser Ser TCG TCA TCT TCC AGT AGC 4.31 11.44 15.70 17.92 12.25 19.54 /1000 frac 0.05 0.14 0.19 0.22 0.15 0.24 Pro Pro Pro Pro CCG CCA CCT CCC 6.33 17.10 18.31 18.42 0.11 0.28 0.30 0.31 AA codon /1000 frac 0.40 0.08 0.13 0.20 Leu Leu Leu Leu CTG CTA CTT CTC 39.95 7.89 12.97 20.04 Ala Ala Ala GCG GCA GCT Ala 6.72 0.10 15.80 0.23 20.12 0.29 GCC 26.51 0.38 Gln CAG Gln 34.18 0.75 CAA 11.51 0.25 CODON USAGE AND LIKELIHOOD RATIO An ORF is more “believable” than another if it has more “likely” codons Do sliding window calculations to find ORFs that have the “likely” codon usage Allows for higher precision in identifying true ORFs; much better than merely testing for length. However, average vertebrate exon length is 130 nucleotides, which is often too small to produce reliable peaks in the likelihood ratio Further improvement: in-frame hexamer count (frequencies of pairs of consecutive codons) GENE PREDICTION AND MOTIFS Upstream regions of genes often contain motifs that can be used for gene prediction ATG -35 -10 0 TTCCAA TATACT Pribnow Box 10 GGAGG Ribosomal binding site Transcription start site STOP RIBOSOMAL BINDING SITE CG-ISLANDS Given 4 nucleotides: probability of occurrence is ~ 1/4. Thus, probability of occurrence of a dinucleotide is ~ 1/16. However, the frequencies of dinucleotides in DNA sequences vary widely. In particular, CG is typically underepresented (frequency of CG is typically < 1/16) 34 WHY CG-ISLANDS? CG is the least frequent dinucleotide because C in CG is easily methylated and has the tendency to mutate into T afterwards However, the methylation is suppressed around genes in a genome. So, CG appears at relatively high frequency within these CG islands So, finding the CG islands in a genome is an important problem 35 THANK YOU