* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Recombinant DNA Technology
DNA supercoil wikipedia , lookup
Protein moonlighting wikipedia , lookup
Epigenetics in stem-cell differentiation wikipedia , lookup
RNA interference wikipedia , lookup
Gene expression profiling wikipedia , lookup
Extrachromosomal DNA wikipedia , lookup
Microevolution wikipedia , lookup
Bisulfite sequencing wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Epigenetics wikipedia , lookup
Designer baby wikipedia , lookup
Cancer epigenetics wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
History of genetic engineering wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Point mutation wikipedia , lookup
History of RNA biology wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Polyadenylation wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Helitron (biology) wikipedia , lookup
Nutriepigenomics wikipedia , lookup
RNA silencing wikipedia , lookup
Histone acetyltransferase wikipedia , lookup
Non-coding DNA wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Short interspersed nuclear elements (SINEs) wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Messenger RNA wikipedia , lookup
Transcription factor wikipedia , lookup
Epigenomics wikipedia , lookup
Non-coding RNA wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Epigenetics in learning and memory wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Epitranscriptome wikipedia , lookup
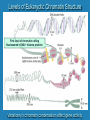
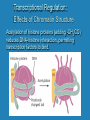
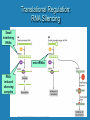
Eukaryotic Gene Expression Managing the Complexities of Controlling Eukaryotic Genes Prokaryotes vs. Eukaryotes Prokaryotes Eukaryotes Closely related genes are clustered together Related genes are located on different chromosomes Largely transcriptional control Significant transcriptional control; other levels of control possible Trans-acting sequencespecific DNA binding proteins Proximal Cis-acting sequences Larger number of larger-sized genes Trans-acting sequencespecific DNA binding proteins Cis-acting sequences can be located at significant distances Control Points for Gene Expression in Eukaryotes DNA Transcriptional Control Post-Transcriptional Control Translational Control transcription RNA translation Protein Post-Translational Control Levels of Eukaryotic Chromatin Structure First level of chromatin coiling Nucleosome = DNA + histone proteins Variations in chromatin condensation affect gene activity. Transcriptional Regulation: Effects of Chromatin Structure Decompaction of chromatin: • Transcription factors unwind nucleosomes in the area where transcription will begin, creating DNAse I hypersensitive sites • RNA polymerase unwinds more nucleosomes as transcription proceeds Transcriptional Regulation: Effects of Chromatin Structure Acetylation of histone proteins (adding -CH3CO) reduces DNA-histone interaction, permitting transcription factors to bind. Transcriptional Regulation: Effects of Chromatin Structure DNA Methylation • DNA Methylation (adding -CH3) can occur on cytosines at CpG groupings near transcription start sites •Inactive genes have methylated cytosines •Active genes have demethylated cytosines • Acetylation of histones is associated with cytosine demethylation Transcriptional Regulation: Control of Initiation •Transcriptional Activator Proteins assist in the formation or action of the basal transcription apparatus Transcriptional Regulation: Control of Initiation • Transcriptional Activator Proteins bind to Enhancer sequences that increase transcription – Enhancers can influence promoters at distances of 50 kb or greater due to DNA looping mechanism – Insulators control the direction of enhancer action Transcriptional Regulation: Control of Initiation • Transcriptional Repressor Proteins have three possible modes of action – compete with activators for DNA binding sites – bind to sites near activator site and inhibit activator contact with basal transcription apparatus – interfere with assembly of basal transcription apparatus Post-Transcriptional Regulation: Alternative RNA Splicing Post-Transcriptional Regulation: RNA Editing Base substitution = after transcription Translational Regulation: RNA Stability • Degradation of mRNA can occur from the 5’ or 3’ end • Stability of mRNA depends on – 5’ cap – 3’ poly-A tail – 5’ and 3’ UTRs: serve as binding sites for regulatory factors – Coding region • Example: Hormone prolactin increases the longevity of casein mRNA coding for milk protein in lactating mammals Translational Regulation • Masking of mRNAs – Many species store mRNAs in the cytoplasm of the egg. These mRNAs are inactive due to masking by proteins. Fertilization of the egg initiates unmasking and translation of these mRNAs. • Availability of specific tRNAs – In the embryonic development of a hornworm, an mRNA is present from day 1 but a specific tRNA needed for its translation is not produced until day 6. Translational Regulation: RNA Silencing Small interfering RNAs microRNAs RNAinduced silencing complex Post-Translational Modification: Phosphorylation Addition or removal of a phosphate group is a common way to change protein activity. Post-Translational Modification: Peptide cleavage Proteins that have an inactive form after synthesis are activated by removal of a small number of amino acids. Prothrombin Cleavage Thrombin Activation of blood clotting factors by cleavage Fibrinogen Cleavage Fibrin Fibrin polymer (blood clot)