Yeast Supplemental Material
... How many generations are too many? This varies from brewery to brewery and strain to strain, but typically yeast can stay vital and genetically stable for 10-30 generations, depending on many factors. If yeast has been stressed for any of the following reasons, it can reduce the viability/vitality o ...
... How many generations are too many? This varies from brewery to brewery and strain to strain, but typically yeast can stay vital and genetically stable for 10-30 generations, depending on many factors. If yeast has been stressed for any of the following reasons, it can reduce the viability/vitality o ...
Gene Section MLL (myeloid/lymphoid or mixed lineage leukemia) Atlas of Genetics and Cytogenetics
... 3969 amino acids; 431 KDa; contains from N-term to C-term 3 AT hooks homologous to high mobility group proteins HMGA1 and HMGA2, binding to the minor grove of DNA; 2 speckled nuclear localisation signals; 2 repression domains RD1 and RD2: RD1 or CXXC: cystein methyl transferase, binds CpG rich DNA, ...
... 3969 amino acids; 431 KDa; contains from N-term to C-term 3 AT hooks homologous to high mobility group proteins HMGA1 and HMGA2, binding to the minor grove of DNA; 2 speckled nuclear localisation signals; 2 repression domains RD1 and RD2: RD1 or CXXC: cystein methyl transferase, binds CpG rich DNA, ...
PDF
... Utx mutant cells proliferate like wild-type cells Previous studies reported that Utx1 cell clones in imaginal discs show a growth advantage relative to neighbouring wild-type cells and proposed that Utx acts as a ...
... Utx mutant cells proliferate like wild-type cells Previous studies reported that Utx1 cell clones in imaginal discs show a growth advantage relative to neighbouring wild-type cells and proposed that Utx acts as a ...
a complex voyage to the X chromosome
... entry site (CES) recognition (Scott et al., 2000; Morales et al., 2004; Li et al., 2005). The coiled coil (CC) domain and PEHE domain (named for its characteristic amino acid composition) are shown (Marin, 2003). (B) The RING finger motif of the MSL2 protein is involved in interactions with MSL1, wh ...
... entry site (CES) recognition (Scott et al., 2000; Morales et al., 2004; Li et al., 2005). The coiled coil (CC) domain and PEHE domain (named for its characteristic amino acid composition) are shown (Marin, 2003). (B) The RING finger motif of the MSL2 protein is involved in interactions with MSL1, wh ...
- Wiley Online Library
... family, which has members present in all kingdoms of life. All SMC proteins appear to regulate chromosome morphology in some way or another. They have been implicated in chromosome condensation, recombination, repair and gene dosage compensation, as well as sister chromatid cohesion. The hallmark of ...
... family, which has members present in all kingdoms of life. All SMC proteins appear to regulate chromosome morphology in some way or another. They have been implicated in chromosome condensation, recombination, repair and gene dosage compensation, as well as sister chromatid cohesion. The hallmark of ...
Cormack et al, 1991 Cell
... Rescue an Otherwise Human TFIID The experiments described in Figure 3 define regions of yeast TFIID core that cannot be replaced by the homologous region of human TFIID to generate a fully functional protein. To determine if the same regions of yeast TFIID could supply the essential functions to an ...
... Rescue an Otherwise Human TFIID The experiments described in Figure 3 define regions of yeast TFIID core that cannot be replaced by the homologous region of human TFIID to generate a fully functional protein. To determine if the same regions of yeast TFIID could supply the essential functions to an ...
Relationships Between RNA Polymerase II Activity and Spt
... yeast genome and many of these exhibit modulation by many factors (Martens et al. 2004, 2005; Hainer et al. 2011; Bird et al. 2006). Furthermore, widespread antisense transcription is being revealed as a mechanism for shaping gene regulation in a number of ways, including transcription over promoter ...
... yeast genome and many of these exhibit modulation by many factors (Martens et al. 2004, 2005; Hainer et al. 2011; Bird et al. 2006). Furthermore, widespread antisense transcription is being revealed as a mechanism for shaping gene regulation in a number of ways, including transcription over promoter ...
invited review
... Nuclear receptors, such as VDR, are characterized by a highly conserved DBD, which is formed by 2 zinc fingers specifically contacting the sequence RGKTSA (R = A or G, K = G or T, S = C or G) within the major groove of genomic DNA (Shaffer and Gewirth 2002) (Fig. 1A). Like most other transcription fac ...
... Nuclear receptors, such as VDR, are characterized by a highly conserved DBD, which is formed by 2 zinc fingers specifically contacting the sequence RGKTSA (R = A or G, K = G or T, S = C or G) within the major groove of genomic DNA (Shaffer and Gewirth 2002) (Fig. 1A). Like most other transcription fac ...
Sampson, Probing the Specifics of Substrate Binding for
... present in COX and the exceedingly complex nature of the process, it was not practical to use the entire enzyme for the minimization study: Instead, as done by Roberts and Pique (23), we used only a fraction of the enzyme. A region of ~25 Å in radius which encompassed all of the amino acid residues ...
... present in COX and the exceedingly complex nature of the process, it was not practical to use the entire enzyme for the minimization study: Instead, as done by Roberts and Pique (23), we used only a fraction of the enzyme. A region of ~25 Å in radius which encompassed all of the amino acid residues ...
Ph1
... • 1952 – became clear that the corresponding chromosomes of the three different genomes are genetically very closely related • Riley and Chapman (1958) - discovered that homoeologous pairing is suppressed by a gene or genes on the long arm of ...
... • 1952 – became clear that the corresponding chromosomes of the three different genomes are genetically very closely related • Riley and Chapman (1958) - discovered that homoeologous pairing is suppressed by a gene or genes on the long arm of ...
Treble clef finger—a functionally diverse zinc
... halves of the zinc-binding site display distinct patterns of sequence conservation. The sequence signal in the N-terminal sub-site is characteristic of a classical zinc knuckle (CPXCG consensus) with small residues immediately following the cysteines. Glycine followed by a primarily hydrophilic resi ...
... halves of the zinc-binding site display distinct patterns of sequence conservation. The sequence signal in the N-terminal sub-site is characteristic of a classical zinc knuckle (CPXCG consensus) with small residues immediately following the cysteines. Glycine followed by a primarily hydrophilic resi ...
The Rad50 Signature Motif: Essential to ATP Binding and
... manganese ions for catalysis and form a DNAbinding groove that can accommodate a doublestranded DNA end.10 In vitro, Mre11 homologs exhibit 30 to 50 exonuclease activity on DNA substrates, as well as endonuclease activity on constrained structures such as hairpin ends.11 – 16 In every organism studi ...
... manganese ions for catalysis and form a DNAbinding groove that can accommodate a doublestranded DNA end.10 In vitro, Mre11 homologs exhibit 30 to 50 exonuclease activity on DNA substrates, as well as endonuclease activity on constrained structures such as hairpin ends.11 – 16 In every organism studi ...
Splicing regulation: a structural biology perspective
... RNP (ribonucleoprotein domain) is the most abundant RNA-binding domain in higher vertebrates (this motif is present in about 0.5%-1% of human genes) [6]. Over the last ten years, biochemical and structural studies have shown that this domain is not only involved in RNA/DNA recognition but also in pr ...
... RNP (ribonucleoprotein domain) is the most abundant RNA-binding domain in higher vertebrates (this motif is present in about 0.5%-1% of human genes) [6]. Over the last ten years, biochemical and structural studies have shown that this domain is not only involved in RNA/DNA recognition but also in pr ...
Bonus, a Drosophila TIF1 homologue, is a chromatin
... variegation of the w+ gene, suggesting that Bon is playing important positive and negative regulatory role in transcription. Finally, chromatin immunoprecipitation assays indicate that Bon is associated with the histone cluster and Stellate locus, two loci that display properties of β−heterochromati ...
... variegation of the w+ gene, suggesting that Bon is playing important positive and negative regulatory role in transcription. Finally, chromatin immunoprecipitation assays indicate that Bon is associated with the histone cluster and Stellate locus, two loci that display properties of β−heterochromati ...
Morphology of nuclear transcription | SpringerLink
... three-dimensional folding of chromatin (maintained by insulators), which place regulatory elements in the same nuclear compartment (Gavrilov et al. 2013; Kosak and Groudine 2004). Other factors were also implicated in loop formation. Using genome-wide ChIP-Seq, it was found that the chromatin remode ...
... three-dimensional folding of chromatin (maintained by insulators), which place regulatory elements in the same nuclear compartment (Gavrilov et al. 2013; Kosak and Groudine 2004). Other factors were also implicated in loop formation. Using genome-wide ChIP-Seq, it was found that the chromatin remode ...
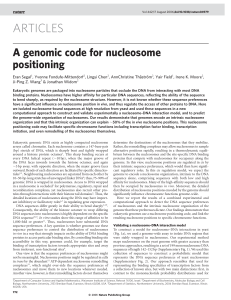
ARTICLES - Weizmann Institute of Science
... for histone–DNA association3); second, when constructing the model we represent the two-fold symmetry axis of the nucleosome structure1 by including the reverse complement of each sequence in the nucleosome collection. More sophisticated nucleosome–DNA interaction models based on mixture models15 or ...
... for histone–DNA association3); second, when constructing the model we represent the two-fold symmetry axis of the nucleosome structure1 by including the reverse complement of each sequence in the nucleosome collection. More sophisticated nucleosome–DNA interaction models based on mixture models15 or ...
Biophysics 101 Genomics and Computational Biology
... Isolation of altered specificity mutants of the single-chain 434 repressor that recognize asymmetric DNA sequences containing TTAA Mechanisms of spontaneous mutagenesis: clues from altered mutational specificity in DNA repair-defective strains. Molecular basis of altered enzyme specificities in a fa ...
... Isolation of altered specificity mutants of the single-chain 434 repressor that recognize asymmetric DNA sequences containing TTAA Mechanisms of spontaneous mutagenesis: clues from altered mutational specificity in DNA repair-defective strains. Molecular basis of altered enzyme specificities in a fa ...
Identification of functional domains in Arabidopsis thaliana mRNA
... Nucleic Acids Research, 2008, Vol. 36, No. 1 203–216 ...
... Nucleic Acids Research, 2008, Vol. 36, No. 1 203–216 ...
Gene Section USF1 (upstream transcription factor 1) Atlas of Genetics and Cytogenetics
... crucial role in chromatin barrier insulator function, in which euchromatin regions are protected from heterochromatin-induced gene silencing (Huang et al., 2007). USFs recruit histone modifying enzymes to the insulator element, which modify the adjacent nucleosomes thereby maintaining chromatin in a ...
... crucial role in chromatin barrier insulator function, in which euchromatin regions are protected from heterochromatin-induced gene silencing (Huang et al., 2007). USFs recruit histone modifying enzymes to the insulator element, which modify the adjacent nucleosomes thereby maintaining chromatin in a ...
Assessment by Molecular Dynamics Simulations of the Structural
... EGR1 reveals a tandem array of three Cys2-His2 zinc finger domains in their C-terminal regions (Figure 1). Structurally, a Cys2-His2 zinc finger is a bba motif stabilized by chelation of a zinc ion between the thiolate groups of a pair of cysteine residues from the b-sheet and the imidazole rings of ...
... EGR1 reveals a tandem array of three Cys2-His2 zinc finger domains in their C-terminal regions (Figure 1). Structurally, a Cys2-His2 zinc finger is a bba motif stabilized by chelation of a zinc ion between the thiolate groups of a pair of cysteine residues from the b-sheet and the imidazole rings of ...
The presence of two UvrB subunits in the UvrAB complex ensures
... size of a UvrA2B1±DNA complex (296 kDa), but also that of a UvrA2B2±DNA complex (372 kDa). As shown above for UvrB±DNA complexes, the DNA wrapped around the UvrB protein also contributes to the size of the complex, thereby increasing the measured volume over that expected based on proteins alone. Mo ...
... size of a UvrA2B1±DNA complex (296 kDa), but also that of a UvrA2B2±DNA complex (372 kDa). As shown above for UvrB±DNA complexes, the DNA wrapped around the UvrB protein also contributes to the size of the complex, thereby increasing the measured volume over that expected based on proteins alone. Mo ...
Clamp loader structure predicts the architecture of DNA polymerase
... structure? The β interactive element site on the δ subunit is contained within the amino-terminal domain (see Figure 3a). The β interactive element on δ is fairly exposed in the γ3δδ′ structure, and appears available to bind β. From the δ–β crystal structure, one can attempt to dock β onto δ in the ...
... structure? The β interactive element site on the δ subunit is contained within the amino-terminal domain (see Figure 3a). The β interactive element on δ is fairly exposed in the γ3δδ′ structure, and appears available to bind β. From the δ–β crystal structure, one can attempt to dock β onto δ in the ...
Mutations of the PC2 Substrate Binding Pocket Alter Enzyme
... of a prodomain, believed to assist in correct folding of the enzyme (2– 4), a catalytic domain with some homology to bacterial subtilisin, and a P- or homo-B domain that plays an essential role in regulating stability and modulating activity (4). The proprotein convertases are thought to be responsi ...
... of a prodomain, believed to assist in correct folding of the enzyme (2– 4), a catalytic domain with some homology to bacterial subtilisin, and a P- or homo-B domain that plays an essential role in regulating stability and modulating activity (4). The proprotein convertases are thought to be responsi ...
Wk12-DeanApop
... The COOH-terminal Region of HBx Is Sufficient to Induce Transcriptional Activity of Egr • Amino terminus did not activate transcription, but carboxyl terminus did (Fig. 8B) • Egr-3 expression was increased by carboxyl terminus, but not by amino terminus (Fig. 8C) ...
... The COOH-terminal Region of HBx Is Sufficient to Induce Transcriptional Activity of Egr • Amino terminus did not activate transcription, but carboxyl terminus did (Fig. 8B) • Egr-3 expression was increased by carboxyl terminus, but not by amino terminus (Fig. 8C) ...
The structure of RNase E at the core of the RNA
... studies using X-ray spectroscopy13. In agreement with our mutagenic studies, the zinc is coordinated at the intra-domain linker through a pair of cysteine residues in the conserved CPxCxGxG motif that occurs throughout the RNase E/RNase G family13 (Fig. S1). A similar motif is found in the metal co ...
... studies using X-ray spectroscopy13. In agreement with our mutagenic studies, the zinc is coordinated at the intra-domain linker through a pair of cysteine residues in the conserved CPxCxGxG motif that occurs throughout the RNase E/RNase G family13 (Fig. S1). A similar motif is found in the metal co ...
Histone acetyltransferase

Histone acetyltransferases (HATs) are enzymes that acetylate conserved lysine amino acids on histone proteins by transferring an acetyl group from acetyl CoA to form ε-N-acetyllysine. DNA is wrapped around histones, and, by transferring an acetyl group to the histones, genes can be turned on and off. In general, histone acetylation increases gene expression.In general, histone acetylation is linked to transcriptional activation and associated with euchromatin. When it was first discovered, it was thought that acetylation of lysine neutralizes the positive charge normally present, thus reducing affinity between histone and (negatively charged) DNA, which renders DNA more accessible to transcription factors. Research has emerged, since, to show that lysine acetylation and other posttranslational modifications of histones generate binding sites for specific protein–protein interaction domains, such as the acetyllysine-binding bromodomain. Histone acetyltransferases can also acetylate non-histone proteins, such as nuclear receptors and other transcription factors to facilitate gene expression.