* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Molecular Biology of the Cell
Mitochondrial DNA wikipedia , lookup
DNA polymerase wikipedia , lookup
Minimal genome wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Genomic imprinting wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Y chromosome wikipedia , lookup
Bisulfite sequencing wikipedia , lookup
Gel electrophoresis of nucleic acids wikipedia , lookup
Comparative genomic hybridization wikipedia , lookup
Genome (book) wikipedia , lookup
United Kingdom National DNA Database wikipedia , lookup
DNA damage theory of aging wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Genome evolution wikipedia , lookup
Human genome wikipedia , lookup
DNA vaccination wikipedia , lookup
Point mutation wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Genealogical DNA test wikipedia , lookup
Molecular cloning wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Epigenetics in stem-cell differentiation wikipedia , lookup
Primary transcript wikipedia , lookup
Cell-free fetal DNA wikipedia , lookup
Genomic library wikipedia , lookup
Epigenetics wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Histone acetyltransferase wikipedia , lookup
Designer baby wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
X-inactivation wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Nucleic acid double helix wikipedia , lookup
Epigenetics in learning and memory wikipedia , lookup
Cancer epigenetics wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
DNA supercoil wikipedia , lookup
Non-coding DNA wikipedia , lookup
Microevolution wikipedia , lookup
History of genetic engineering wikipedia , lookup
Extrachromosomal DNA wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Helitron (biology) wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Neocentromere wikipedia , lookup
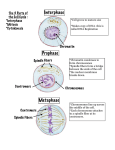
TIGP Molecular and Cellular Biology DNA, Chromosomes and Genome (Chapter 4) Textbook: Molecular Biology of the Cell Alberts et al. Sixth Edition Instructor: Chung-Ju Rachel Wang Sept 21, 2015 Outlines • • • • THE STRUCTURE AND FUNCTION OF DNA CHROMOSOMAL DNA AND ITS PACKAGING IN THE CHROMATIN FIBER CHROMATIN STRUCTURE AND FUNCTION THE GLOBAL STRUCTURE OF CHROMOSOMES Introduction Genetic material is packed in a set of chromosomes Introduction THE STRUCTURE AND FUNCTION OF DNA Watson and Crick solved the puzzle of DNA structure by x-ray diffraction (to determine the 3D atomic structure). The model was proposed in 1953, that nicely explains how DNA can be replicated and inherited as the genetic material. A DNA Molecule Consists of Two Complementary Chains of Nucleotides Polarity: 5’->3’ Antiparallel A DNA Molecule Consists of Two Complementary Chains of Nucleotides Sequence of nucleotides on each strand must be complementary Two hydrogen bonds A, G: purine T, C: pyrimidine Three hydrogen bonds A DNA Molecule Consists of Two Complementary Chains of Nucleotides the double helix is right-handed with about 10.4 nucleotides per turn THE STRUCTURE AND FUNCTION OF DNA • The Structure of DNA Provides a Mechanism for Heredity The Structure of DNA Provides a Mechanism for Heredity DNA contains a sequence of nucleotides, like the letters of a document written in an alphabetic script. Each strand can acts as a template for the synthesis of a new complementary strand. The Structure of DNA Provides a Mechanism for Heredity The nucleotide sequence of the human beta-globin gene. The DNA highlighted in yellow show the three regions of the gene that specify the amino acid sequence for the beta-globin protein. The complete store of information in an organism’s DNA is called its genome. Human genome contains 3.2 billion nucleotide pairs. THE STRUCTURE AND FUNCTION OF DNA • In Eukaryotes, DNA Is Enclosed in a Cell Nucleus In Eukaryotes, DNA Is Enclosed in a Cell Nucleus Human fibroblast Two lipid bilayer membranes DNA in a eukaryotic cell is sequestered in a nucleus, a compartment is encircled by nuclear envelope. CHROMOSOMAL DNA AND ITS PACKAGING IN THE CHROMATIN FIBER • Eukaryotic DNA Is Packaged into a Set of Chromosomes Human genome (1C): 3.2 x 109 bp 6.4 X 109 bp DNA is distributed in 46 different chromosomes. If DNA in a single human cell can be laid end-to-end, they would reach 2 meters. How 2 meters of DNA can fit inside a small nucleus which is only 6 µm in diameter (equivalent to pack 40km thread in a tennis ball. ) 40 Km thread Eukaryotic DNA Is Packaged into a Set of Chromosomes Chromosomes Contain Long Strings of Genes Two sets of chromosomes: one from father and one from mother “Chromosome painting” technique by DNA hybridization can distinguish each pair of chromosomes. Mitosis 23 pairs of homologous chromosomes Each chromosome consists of a single, long linear DNA molecular along with proteins that fold and pack the DNA into a compact structure. Eukaryotic DNA Is Packaged into a Set of Chromosomes Chromosomes Contain Long Strings of Genes Karyotype by Giemsa staining. centromere The chromosomes for this staining are at an early stage in mitosis, when the chromosomes are incompletely compacted. Red knobs indicate positions of genes that code for the large ribosomal RNAs. The banding patterns reflect variations in chromatin structure. The band pattern on each chromosome is unique, providing the initial means to identify and number each chromosome. Eukaryotic DNA Is Packaged into a Set of Chromosomes Chromosomes Contain Long Strings of Genes Aberrant human chromosomes: In an individual carrying a balanced chromosomal translocation, the chromosome painting technique can be used to detect these chromosomal rearrangement. CHROMOSOMAL DNA AND ITS PACKAGING IN THE CHROMATIN FIBER • Chromosomes Contain Long Strings of Genes Chromosomes Contain Long Strings of Genes A gene is defined as a segment of DNA that contains the instructions for making a particular protein or even RNA molecule as their final product. 16 chromosomes, high density of genes. Many genomes of multicellular organisms contain, in addition to genes, a large quantity of interspersed DNA. Chromosomes Contain Long Strings of Genes Two closely related species of deer with very different chromosome numbers. There is no simple relationship between chromosome number, complexity of the organism, and total genome size. CHROMOSOMAL DNA AND ITS PACKAGING IN THE CHROMATIN FIBER • The Nucleotide Sequence of the Human Genome Shows How Our Genes Are Arranged The Nucleotide Sequence of the Human Genome Shows How Our Genes Are Arranged Dark brown: known genes Red: predicted genes The Nucleotide Sequence of the Human Genome Shows How Our Genes Are Arranged About 1300 bp encode a protein. Little of the genome codes for proteins CHROMOSOMAL DNA AND ITS PACKAGING IN THE CHROMATIN FIBER • Each DNA Molecule That Forms a Linear Chromosome Must Contain a Centromere, Two Telomeres, and Replication Origins Each DNA Molecule That Forms a Linear Chromosome Must Contain a Centromere, Two Telomeres, and Replication Origins During interphase, the cell is actively expressing its genes and is therefore synthesizing proteins. Also, DNA is replicated to produce two closely paired sister DNA molecules. At M phase, chromosomes condense, the nuclear envelope breaks down, the mitotic spindle forms to separate sister chromosomes. Sister chromatids Each DNA Molecule That Forms a Linear Chromosome Must Contain a Centromere, Two Telomeres, and Replication Origins During interphase, chromatin exists as long threads, so that individual chromosomes cannot be distinguished. The highly condensed chromosomes at M phase are known as mitotic chromosomes. Most easily visualized. Each chromosome operates as a distinct structural unit, and it contains three specialized components for its functions: Replication origin centromere, Telomeres. Each DNA Molecule That Forms a Linear Chromosome Must Contain a Centromere, Two Telomeres, and Replication Origins CHROMOSOMAL DNA AND ITS PACKAGING IN THE CHROMATIN FIBER • DNA Molecules Are Highly Condensed in Chromosomes • Nucleosomes Are a Basic Unit of Eukaryotic Chromosome Structure Nucleosomes Are a Basic Unit of Eukaryotic Chromosome Structure The proteins that bind to DNA to form chromosomes are traditionally divided into histones and non-histone chromosomal proteins. Nucleosomes Are a Basic Unit of Eukaryotic Chromosome Structure Nucleosomes as seen in the electron microscope. Nucleosome discovered in 1974. A human cell with 6.4 x 109 bp contains ~30 M nucleosomes. A. Chromatin isolated from an interphase nucleus appears as a thread about 30nm thick. B. The unpacked chromatin (beads on string) with nucleosomes and their linker DNA. Nucleosomes Are a Basic Unit of Eukaryotic Chromosome Structure After digested by nucleases that degrade linker DNA, each nucleosome core particle consists of 1 complex of 8 histone proteins and 147 nt DNA. The linker DNA that separates each nucleosome core vary in length from a few bp to about 80. On average, nucleosome repeat at intervals about 200 bp. CHROMOSOMAL DNA AND ITS PACKAGING IN THE CHROMATIN FIBER • The Structure of the Nucleosome Core Particle Reveals How DNA Is Packaged The Structure of the Nucleosome Core Particle Reveals How DNA Is Packaged A disc-shaped histone core with DNA tightly wrapped in a left-handed coil of 1.7 turns. DNA in gray The Structure of the Nucleosome Core Particle Reveals How DNA Is Packaged • Very conserved, small proteins (102-135 aa) • Share a structural motif (histone fold), with three alpha- helices and two loops. H3-H4 and H2A-H2B form heterodimers. • C. H2A and 2B form dimer Each core histone contains an N-tail, which is subjected to several forms of modification, and a histone fold region. N-terminal tails protrude from the disc-shaped core structure, which is highly flexible. The Structure of the Nucleosome Core Particle Reveals How DNA Is Packaged 142 hydrogen bonds are formed between DNA and histone core, , providing the tight association. More than 1/5 of the amino acid of core histones are either lysine or arginine, and their positive charges can effectively neutralize the negatively charged DNA backbone. The minor groove is compressed on the inside of the turn. CHROMOSOMAL DNA AND ITS PACKAGING IN THE CHROMATIN FIBER • Nucleosomes Have a Dynamic Structure, and Are Frequently Subjected to Changes Catalyzed by ATP-Dependent Chromatin Remodeling Complexes Nucleosomes Have a Dynamic Structure, and Are Frequently Subjected to Changes Catalyzed by ATP-Dependent Chromatin Remodeling Complexes A. Using the energy of ATP hydrolysis, the remodeling complex can push on the DNA and loosen its attachment to the nucleosome core. Each cycle of ATP binding, ATP hydrolysis, and release of the ADP and Pi products moves the DNA on histone core. B. The structure of a nucleosome-bound dimer of two ATPase subunits (green) that slide nucleosomes in the ISW1 family of chromatin remodeling complexes. C. Another large chromatin remodeling complex contains 15 units, including an ATPase. Hydrolyzes ATP to catalyze nucleosome sliding by pulling the nucleosome core along the DNA, that makes specific DNA sequences available to other proteins. Schematics showing two mechanisms for a dinucleosome spacing reaction. In scheme (1), the linker DNA is flexible, and in scheme (2), the protein chain between HAND and the ATPase domain is flexible. K Yamada et al. Nature 472, 448-453 (2011) doi:10.1038/nature09947 Nucleosomes Have a Dynamic Structure, and Are Frequently Subjected to Changes Catalyzed by ATP-Dependent Chromatin Remodeling Complexes •Cells contain dozens of different chromatin remodeling complex, that consists of 10 or more subunits. This process is carefully controlled. As genes are turned on and off, chromatin remodeling complexes are brought to specific regions of DNA where they act locally to influence chromatin structure. Nucleosome removal and histone exchange catalyzed by ATP-dependent chromatin remodeling complexes and histone chaperones. CHROMOSOMAL DNA AND ITS PACKAGING IN THE CHROMATIN FIBER • Nucleosomes Are Usually Packed Together into a Compact Chromatin Fiber Nucleosomes Are Usually Packed Together into a Compact Chromatin Fiber How nucleosome are organized into condensed arrays is still not clear. A. zigzag model from x-ray crystallography. The conformation of histones in a tetranucleosome. It is a possible mode for the 30-nm chromatin fiber. Nucleosomes Are Usually Packed Together into a Compact Chromatin Fiber Histone tails may help to pack nucleosome together into 30 nm fiber in which the tails of one nucleosome contact the histone core of an adjacent nucleosome. Nucleosomes Are Usually Packed Together into a Compact Chromatin Fiber The position and structure of histone H1 is shown. The H1 constrains about additional 20 bp of DNA. CHROMATIN STRUCTURE AND FUNCTION • Heterochromatin Is Highly Organized and Restricts Gene Expression • The Heterochromatic State Is Self-Propagating Genetic inheritance and epigenetic inheritance • Chromatin structure plays a central role in the development and growth. Cell memory Epigenetic information is usually erased during the formation of eggs and sperm. 45 Heterochromatin is highly organized and resistant to gene expression Heterochromatin (highly condensed form) Genes there are silenced. 46 The Heterochromatic State Is Self-Propagating The cause of position effect variegation in fly. Heterochromatin (green) is normally prevented from spreading into adjacent regions of euchromatin (red) by barrier DNA sequences. In this case, the barrier DNA is removed, heterochromatin can spread into neighboring DNA, proceeding for different distances in different cells. The Heterochromatic State Is Self-Propagating The wild-type White+ gene is moved near heterochromatin, and White gene’s expression is affected by nearby heterochromatin. The white- patches represent cell lineages in which the White+ gene has been silenced. CHROMATIN STRUCTURE AND FUNCTION • The Core Histones Are Covalently Modified at Many Different Sites The Core Histones Are Covalently Modified at Many Different Sites The Core Histones Are Covalently Modified at Many Different Sites The Core Histones Are Covalently Modified at Many Different Sites •These modifications are reversible. •Added or removed by enzymes. •Each enzyme is recruited to specific sites on the chromatin at defined times. •Can further attract specific proteins for various biological functions, for example, determining how and when genes will be expressed. CHROMATIN STRUCTURE AND FUNCTION • Chromatin Acquires Additional Variety Through the Site-Specific Insertion of a Small Set of Histone Variants Chromatin Acquires Additional Variety Through the Site-Specific Insertion of a Small Set of Histone Variants These histone variants can replace the regular histones by ATP-dependent chromatin remodeling enzymes and histone chaperones. CHROMATIN STRUCTURE AND FUNCTION • Covalent Modifications and Histone Variants Act in Concert to Control Chromosome Functions Covalent Modifications and Histone Variants Act in Concert to Control Chromosome Functions PHD domain (gray) binds to histone tail (green) H3 lysine 4trimethylation. B. space-filling model of an ING PHD domain bound to a histone tail. C. A ribbon model showing how the N-terminal six amino acids in the H3 tail are recognized. Red lines represent hydrogen bonds. Covalent Modifications and Histone Variants Act in Concert to Control Chromosome Functions The covalent modifications and histone variants act in concert to produce a “histone code” that helps to determine biological function. The reader complex can bind tightly only to a region of chromatin that contains several of different histone marks. Particular combinations of markings on chromatin to attract additional protein complexes that execute biological function at the right time. Covalent Modifications and Histone Variants Act in Concert to Control Chromosome Functions Methylation and acetylation on the same residue can not exist at the same time. CHROMATIN STRUCTURE AND FUNCTION • A Complex of Reader and Writer Proteins Can Spread Specific Chromatin Modifications Along a Chromosome A Complex of Reader and Writer Proteins Can Spread Specific Chromatin Modifications Along a Chromosome How the recruitment of a reader-writer complex can spread chromatin changes along a chromosome. The writer is an enzyme that creates a specific modification on histone. After its recruitment to a specific site on a chromosome by a transcriptional regulatory protein, the writer collaborate with a reader protein to spread its mark. CHROMATIN STRUCTURE AND FUNCTION • Barrier DNA Sequences Block the Spread of Reader–Writer Complexes and thereby Separate Neighboring Chromatin Domains Barrier DNA Sequences Block the Spread of Reader–Writer Complexes and thereby Separate Neighboring Chromatin Domains (A) The tethering of a region of chromatin to nuclear pore complex. (B) The tight binding of barrier proteins to a group of nucleosomes. (C) By recruiting a histone modifying enzymes, barriers can erase the histone marks that are required for heterochromatin to spread. For example, HS4 barrier DNA separates the active chromatin domain that contains the beta-globin gene from an adjacent region (16-Kb) of silenced condensed chromatin. Deletion of HS4 sequence causes poor expression of globin genes. HS4 sequence contains a cluster of binding sites for histone acetyltransferases. CHROMATIN STRUCTURE AND FUNCTION • The Chromatin in Centromeres Reveals How Histone Variants Can Create Special Structures The Chromatin in Centromeres Reveals How Histone Variants Can Create Special Structures In budding yeast, a special centromeric DNA sequence assembles a single nucleosome in which two copies of an H3 variant histone (CENP-A) replace the normal H3. The yeast centromere form kinetochore to capture microtubule. The Chromatin in Centromeres Reveals How Histone Variants Can Create Special Structures Active centromere has CENP-A histone H3 variant epigenetic mark. Human centromeres have plasticity. A series of A-T rich alpha satellite DNA sequences is repeated many thousands of times at each human centromere (red), and is surrounded by pericentric heterochromatin (brown). An ancient chromosome rearrangement results in two blocks of alpha satellite DNA. Only one is stably functional. CHROMATIN STRUCTURE AND FUNCTION • Some Chromatin Structures Can Be Directly Inherited Some Chromatin Structures Can Be Directly Inherited In this model, some of the histone with modifications are distributed to each sister chromosome after DNA replication. CHROMATIN STRUCTURE AND FUNCTION • Experiments with Frog Embryos Suggest that both Activating and Repressive Chromatin Structures Can Be Inherited Epigenetically Experiments with Frog Embryos Suggest that both Activating and Repressive Chromatin Structures Can Be Inherited Epigenetically The well-characterized MyoD gene encoded a master transcriptional regulatory protein for muscular gene promoters, including itself. The active chromatin surrounding the MyoD promoter contains the variant histone H3.3 in a Lys 4 methylate form. By injecting excess mRNA encoding normal H3.3 or mutant H3.3, experiments provide evidence of a gene-activating chromatin state through epigenetic control. CHROMATIN STRUCTURE AND FUNCTION • Chromatin Structures Are Important for Eukaryotic Chromosome Function In a complex multicellular organism, the cells in different lineages must specialize by changing the accessibility and activity of many hundreds of genes. Each cell holds a record of its past developmental history in the regulatory circuits that control its many genes. That record, it seems, is partly stored in the structure of the chromatin. THE GLOBAL STRUCTURE OF CHROMOSOMES • Chromosomes Are Folded into Large Loops of Chromatin Chromosomes Are Folded into Large Loops of Chromatin Lampbrush chromosomes in an amphibian oocyte. •A special form of chromosomes, found in oocytes of most animals (except mammals). •Seen at early meiosis stage due to an active transcription of many genes. •DNA are organized into a series of large chromatin loops, emanating from a chromosomal axis. Early in oocyte differentiation, each chromosome replicates to begin meiosis, and chromosomes form this highly extended structure containing a total of four replicated chromatids. Chromosomes Are Folded into Large Loops of Chromatin •The genes on the loop are actively expressed. •The genes condensed close to axis are not expressed. •A given loop always contains the same DNA sequences Chromosome axis Chromosomes Are Folded into Large Loops of Chromatin Chromosome conformation capture (3C) can determine the position of loops in interphase chromosomes. Other methods developed based on 3C 75 Chromosomes Are Folded into Large Loops of Chromatin Each loop contains 50-200k bp DNA THE GLOBAL STRUCTURE OF CHROMOSOMES • Polytene Chromosomes Are Uniquely Useful for Visualizing Chromatin Structures Polytene Chromosomes Are Uniquely Useful for Visualizing Chromatin Structures •Some insect cells, for example salivary gland cells in fly larvae have polytene chromosomes. •All homologous chromosome copies are held side by side, linked together by chromocenter. •Each polytene chromosome contains a thousand identical DNA sequences with dark and light bands with different degree of chromatin condensation. THE GLOBAL STRUCTURE OF CHROMOSOMES • There Are Multiple Forms of Chromatin • Chromatin Loops Decondense When the Genes Within Them Are Expressed Chromatin Loops Decondense When the Genes Within Them Are Expressed H3Lys 9 di-methylation H3 Lys 9 acetylation (green) chromocenter. Pericentric heterochromatin. Marks active genes Decondensed portion of chromosomes is undergoing RNA synthesis. Chromatin Loops Decondense When the Genes Within Them Are Expressed Two homologous chromosome are not in general co-located. Chromatin Loops Decondense When the Genes Within Them Are Expressed •Gene-rich regions are visualized with a FISH probe that hybridizes to the Alu interspersed repeat, which is present in more than a million copies in the human genome. •These sequences cluster in gene-rich regions. •Heterochromatin regions often are associated with the nuclear lamina. Alu signals (green) Chromatin Loops Decondense When the Genes Within Them Are Expressed THE GLOBAL STRUCTURE OF CHROMOSOMES • Chromatin Can Move to Specific Sites Within the Nucleus to Alter Gene Expression Chromatin Can Move to Specific Sites Within the Nucleus to Alter Gene Expression •The position of a gene in the interior of the nucleus changes when it becomes highly expressed. THE GLOBAL STRUCTURE OF CHROMOSOMES • Networks of Macromolecules Form a Set of Distinct Biochemical Environments inside the Nucleus Nuclear subcompartments: Nucleolus rRNA expression and ribosome assembly and maturation as well as many other specialized reactions. Networks of Macromolecules Form a Set of Distinct Biochemical Environments inside the Nucleus Cajal body: 0.3-1 µm in nucleus, may related to RNA-related metabolic processes. These structures without membrane are composed of selected protein and RNA that bind together to create networks for multiple biological functions. Networks of Macromolecules Form a Set of Distinct Biochemical Environments inside the Nucleus THE GLOBAL STRUCTURE OF CHROMOSOMES • Mitotic Chromosomes Are Especially Highly Condensed Mitotic Chromosomes Are Especially Highly Condensed