* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download rec07
Epitranscriptome wikipedia , lookup
Biology and consumer behaviour wikipedia , lookup
Protein moonlighting wikipedia , lookup
Frameshift mutation wikipedia , lookup
Copy-number variation wikipedia , lookup
Genomic imprinting wikipedia , lookup
Genomic library wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Epigenetics in learning and memory wikipedia , lookup
Pathogenomics wikipedia , lookup
Ridge (biology) wikipedia , lookup
Epigenomics wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Non-coding RNA wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Cancer epigenetics wikipedia , lookup
Gene therapy wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Short interspersed nuclear elements (SINEs) wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Transposable element wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Genetic code wikipedia , lookup
Genetic engineering wikipedia , lookup
Gene expression programming wikipedia , lookup
Gene nomenclature wikipedia , lookup
Human genome wikipedia , lookup
Minimal genome wikipedia , lookup
Genome (book) wikipedia , lookup
Gene desert wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Point mutation wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Non-coding DNA wikipedia , lookup
Gene expression profiling wikipedia , lookup
Primary transcript wikipedia , lookup
History of genetic engineering wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Genome editing wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Genome evolution wikipedia , lookup
Microevolution wikipedia , lookup
Designer baby wikipedia , lookup
Helitron (biology) wikipedia , lookup
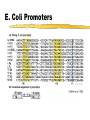
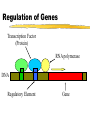
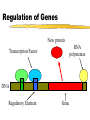
Comp. Genomics Recitation 7 2/4/09 PSSMs+Gene finding Partially based on slides by Irit Gat-Viks and Metsada Pasmanik-Chor Biological Motifs • Biological units with common functions frequently exhibit similarities at the sequence level. These include very short “motifs”, such as: • Gene splice sites • DNA regulatory binding sites (bound by transcription factors) • Often it is desirable to model such motifs, to enable searching for new ones. Probabilistic models are very useful. Today we deal with PSSM - the simplest. E. Coli Promoters Regulation of Genes Transcription Factor (Protein) RNA polymerase (Protein) DNA Regulatory Element Gene Regulation of Genes Transcription Factor (Protein) RNA polymerase DNA Regulatory Element Gene Regulation of Genes New protein RNA polymerase Transcription Factor DNA Regulatory Element Gene Motif Logo • Motifs can mutate on less important bases. Position:1234567 • The five motifs at top right have mutations in position 3 and 5. • Representations called motif logos illustrate the conserved regions of a motif. http://weblogo.berkeley.edu http://fold.stanford.edu/eblocks/acsearch.html TGGGGGA TGAGAGA TGGGGGA TGAGAGA TGAGGGA Example: Calmodulin-Binding Motif (calcium-binding proteins) PSSM Starting Point • A gap-less MSA of known instances of a given motif. Representing the motif by either: • Consensus. • Position Specific Scoring Matrix (PSSM). Usage of a PSSM • For a putative k-mer GTGC– multiply the probabilities: p1(G)·p2(T)·p3(G)·p4(C) • This gives the likelihood of the motif given the PSSM model TATA box motif Gene finding • Only part of the genome encodes proteins • 80-90% in bacteria, ab. 2% in humans • Goal: Given a genome sequence, identify gene boundaries The genetic code • A protein-coding gene, an open reading frame (ORF) begins with an ATG and ends with one of three stop codons Prokaryotic genes • • • • The ‘easy’ problem Difficulty – not all possible ORFs are actually genes In E.Coli: 6500 ORFs while there are 4290 genes. Additional “handles” are needed Handle #1: Long ORFs • In random DNA, one stop codon every 64/3=21 codons on average. • Average protein is ~300 codons long. • => search long ORFs. • Problems: • Short genes • Overlapping long ORFs on opposite strands Handle #2: Codon frequencies • Coding DNA is not random: • In random DNA, expect Leu : Ala : Trp ratio of 6 : 4 : 1 • In real proteins, 6.9 : 6.5 : 1 • Different frequencies for different species. Using Codon Frequencies/Usage • Assume each codon is independent. • For codon abc calculate frequency f(abc) in coding region. • Given coding sequence a1b1c1,…, an+1bn+1cn+1 • Calculate p1 f a1b1c1 f a2b2c2 ... f anbncn p2 f b1c1a2 f b2c2a3 ... f bncnan 1 p3 f c1a2b2 f c2a3b3 ... f cnan 1bn 1 • The probability that the ith reading frame is the coding region: Pi pi p1 p2 p3 16 Handle #3: G+C content • C+G content (“isochore”) has strong effect on gene density, gene length etc. • < 43% C+G : 62% of genome, 34% of genes • >57% C+G : 3-5% of genome, 28% of genes • Gene density in C+G rich regions is 5 times higher than moderate C+G regions and 10 times higher than rich A+T regions • Amount of intronic DNA is 3 times higher for A+T rich regions. (Both intron length and number). • Etc… Handle #4: Promoter motifs • Transcription depends on regulatory regions. • Common regulatory region – the promoter • RNA polymerase binds tightly to a specific DNA sequence in the promoter Gene prediction programs Scan the sequence in all 6 reading frames: 1. Start and stop codons 2. Long ORF 3. Codon usage 4. GC content 5. Gene features: promotor, terminator, poly A sites, exons and introns, … Frame +1 Frame +2 Frame +3 19 Moving to eukaryotes • Less of the genome is protein coding + introns are a (very) serious headache Eukaryote gene structure • • • • • Gene length: 30kb, coding region: 1-2kb Binding site: ~6bp; ~30bp upstream of TSS Average of 6 exons, 150bp long Huge variance: - dystrophin: 2.4Mb long Blood coagulation factor: 26 exons, 69bp to 3106bp; intron 22 contains another unrelated gene 21 Splicing • Splicing: the removal of the introns. • Performed by complexes called spliceosomes, containing both proteins and snRNA. • The snRNA recognizes the splice sites through RNA-RNA base-pairing • Recognition must be precise: a 1nt error can shift the reading frame making nonsense of its message. • Many genes have alternative splicing which changes the protein created. 22 Splice Sites 23