* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Gene regulation - Department of Plant Sciences
Behavioral epigenetics wikipedia , lookup
Bisulfite sequencing wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Transposable element wikipedia , lookup
Genome evolution wikipedia , lookup
Gene therapy wikipedia , lookup
Messenger RNA wikipedia , lookup
Histone acetyltransferase wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Epigenetics wikipedia , lookup
Genetic engineering wikipedia , lookup
Gene desert wikipedia , lookup
Gene nomenclature wikipedia , lookup
Short interspersed nuclear elements (SINEs) wikipedia , lookup
Gene expression programming wikipedia , lookup
RNA silencing wikipedia , lookup
Gene expression profiling wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Epigenetics in stem-cell differentiation wikipedia , lookup
RNA interference wikipedia , lookup
Cancer epigenetics wikipedia , lookup
Non-coding DNA wikipedia , lookup
Non-coding RNA wikipedia , lookup
Point mutation wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Epigenetics of depression wikipedia , lookup
History of genetic engineering wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Epitranscriptome wikipedia , lookup
Microevolution wikipedia , lookup
Epigenomics wikipedia , lookup
Designer baby wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Mir-92 microRNA precursor family wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Helitron (biology) wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Transcription factor wikipedia , lookup
Epigenetics in learning and memory wikipedia , lookup
Nutriepigenomics wikipedia , lookup
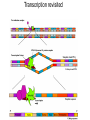
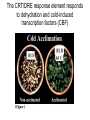
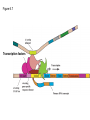
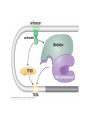
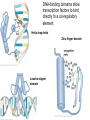
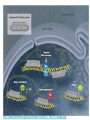
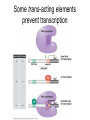
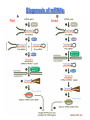
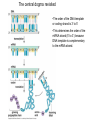
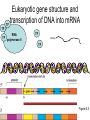
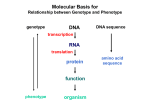
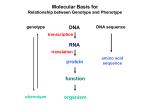
Lecture 9 Chapter 6 Gene expression and regulation II Neal Stewart Focus questions • How important are cis-regulatory elements and trans-acting factors in gene regulation? • What are the control points that can regulate gene expression? Transcription revisited Promoter elements not required for transcription initiation • CAAT box – usually located at -70 to -80 within the promoter • GC box • Other gene-specific elements (lightresponsive, nutrient-responsive, etc.) • Enhancer elements What are some biological roles of transcription factors? • Basal transcription regulation – general transcription factors • Development • Response to intercellular signals • Response to environment • Cell cycle control The CRT/DRE response element responds to dehydration and cold-induced transcription factors (CBF) Figure 6.7 Transcription factors Figure 6.8 Enhancer can work from downstream and upstream region Enhancers • Their location is not fixed. Location could be in the upstream or downstream DNA, in intron, exon or in the untranslated region. • They enhance transcription by acting on promoter in cis (typically) • Each enhancer has its own binding protein. These proteins are trans-regulatory activating factors • Sequence of enhancers is variable. • Enhancers regulate tissue-specific and temporal expression of genes. TATA binding protein (TBP) transcription factor Wikipedia.com DNA-binding domains allow transcription factors to bind directly to a cis-regulatory element Helix-loop-helix Zinc finger domain Leucine zipper domain Extreme trans-acting effectors of transcription: TAL effectors • From plant pathogenic bacteria Xanthomonas • Secreted by bacteria when they infect • Transcriptional activator-like (TAL) effectors bind with plant promoters to express genes beneficial for the bacteria http://www.sciencemag.org/content/333/6051/1843/F2.large.jpg Repression of transcription TFs that act as repressors Some trans-acting elements prevent transcription Introducing RNAi http://www.youtube.com/watch?v= H5udFjWDM3E&feature=related What is a microRNA (miRNA)? Controlling gene expression post-transcriptionally. microRNA is an abundant class of newly identified small non-coding regulatory RNAs. Major characteristics of miRNAs: • 18-26 nt in length with a majority of 21-23 nt • non-coding RNA • derived from a precursor with a long nt sequence • this precursor can form a stem-loop 2nd hairpin structure • the hairpin structure has low minimal free folding energy (MFE) and high MFE index Slide courtesy of Baohong Zhang, East Carolina Univ miRNA regulates plant development miRNA 156 increasing leaf initation, decreasing apical dominance, and forming bushier plant. miRNA 164 stamens are fused together. miRNA 172 sepal and petal disappeared. miRNA 319 Leaf morphology WT miRNA Slide courtesy of Baohong Zhang, East Carolina Univ Small interfering RNAs inhibit expression of a homologous gene Biogenesis of miRNAs Plant Animal Bartel, 2004. Cell. Mechanisms of miRNA-mediated gene regulation Post-transcriptional gene regulation Two major molecular mechanisms Zhang et al. 2006. Developmental Biology Slide courtesy of Baohong Zhang, East Carolina Univ Mary-Dell Chilton • Undergrad and PhD University of Illinois • Postdoc with Gene Nester and Milt Gorgon Univ Washington • One of the first plant transformation Washington University • Career at CibaNovartisSyngenta Pre-transcriptional gene regulation by methylation of DNA and acetylation of histones Special proteins (e.g. chromomethylases) maintain methylation patterns Switching a gene on and off through DNA methylation and histone modification Arabidopsis MET1 Cytosine Methyltransferase Mutants Kankel et al. 2003. 163 (3):1109 Genetics Plants mutant for MET1 show late-flowering phenotypes Histone acetyl transferases and chromatin remodeling allows promoters to be accessible to RNAPII Histone tails are modified and can be studied easily Figure 6.9 Some post-translational modifications • • • • • • • • • Phosphorylation Biotinylation Glycosylation Acetylation Alkylation Methylation Glutamylation Glycylation Isoprenylation • Lipoylation • Phosphopantetheinyl ation • Sulfation • Selenation • C-terminal amidation Phosphorylation is important for intracellular signalling http://www.scq.ubc.ca/wp-content/uploads/2006/07/phosphocascades.gif Protein glycosylation in the ER The central dogma revisited •The order of the DNA template or coding strand is 3’ to 5’ •This determines the order of the mRNA strand (5’ to 3’) because DNA template is complementary to the mRNA strand. Eukaryotic gene structure and transcription of DNA into mRNA TF TF RNA TF polymerase II TF AAAAA TF Figure 6.5 Manipulating gene expression • Can be done at several levels – Promoters, enhancers, transcription factors – Post-transcriptional – Translational – Methylation • Biotechnology typically manipulates promoter • Post-transcriptional gene silencing (RNAi) increasingly important