* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download VII. Some methods for studying gene expression
Genome (book) wikipedia , lookup
Cancer epigenetics wikipedia , lookup
Epigenomics wikipedia , lookup
Protein moonlighting wikipedia , lookup
Genetic code wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Transcription factor wikipedia , lookup
Nucleic acid tertiary structure wikipedia , lookup
Gene therapy wikipedia , lookup
Gene desert wikipedia , lookup
Genome evolution wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
History of genetic engineering wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Epigenetics in learning and memory wikipedia , lookup
Non-coding DNA wikipedia , lookup
Gene nomenclature wikipedia , lookup
Gene expression programming wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Point mutation wikipedia , lookup
Short interspersed nuclear elements (SINEs) wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Mir-92 microRNA precursor family wikipedia , lookup
History of RNA biology wikipedia , lookup
Microevolution wikipedia , lookup
Polyadenylation wikipedia , lookup
Helitron (biology) wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Gene expression profiling wikipedia , lookup
Designer baby wikipedia , lookup
RNA interference wikipedia , lookup
Epigenetics of human development wikipedia , lookup
RNA silencing wikipedia , lookup
Messenger RNA wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Non-coding RNA wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
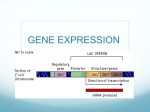
IV. Translation IV. Translation (3) Translation termination i. When a ribosome comes to a nonsense codon (or stop codon, usually one of UAA, UAG and UGA), translation stops and polypeptide is released from ribosome. ii. Stop codons do not encode an amino acid, so they have no corresponding tRNA. iii. Termination requires release factors (RF1 and RF2) which recognize nonsense codon and promote the release of the polypeptide form the tRNA and the ribosome from the mRNA. (Fig. 2.35) Termination of translation at a nonsense codon IV. Translation 5. Polycistronic mRNA ※ In bacteria and archaea, the same mRNA can encode more than one polypeptide. Such mRNAs, called polycistronic mRNAs, must have more than one TIR to allow simultaneous translation of more than one sequence of the mRNA. (1) Even if the two coding regions overlap, the two polypeptides on an mRNA can be translated independently by different ribosomes. (2) Translational coupling – The translation of upstream gene is required for the translation of the gene immediately downstream. The secondary structure of the RNA blocks translation of the second polypeptide unless it is disrupted by a ribosome translating the first coding sequence. Structure of a polycistronic mRNA Even if the two coding regions overlap, the two polypeptides on an mRNA can be translated independently by different ribosomes. IV. Translation (3) Polar effect on gene expression - Some mutations that affect the expression of a gene in a polycistronic mRNA can have secondary effects on the expression of downstream gene. i. The insertion of an transcription terminator prevents the transcription of downstream gene. ii. The mutation changing a codon to a nonsense codon will dissociate the ribosome from mRNA, then the translation of downstream gene that is translationally coupled to the upstream gene will not translated. (4) ρ–dependent polarity (as shown in Fig.2.38) A. Normally the rut site is masked by ribosome translating the mRNA of gene Y. B. If translation is blocked in gene Y by a mutation that changes the codon CAG to UAG, the ρ factor can cause transcription termination before the RNA polymerase reach gene Z. C. Only the fragment of gene Y protein and mRNA are produced and even gene Z is not even transcribed into mRNA. Model for translational coupling in polycistronic mRNA Polarity in transcription of a polycistronic mRNA transcribed from PYZ. V. Regulation of gene expression 1. Transcriptional regulation (1) Genes whose products regulate the expression of other genes are called regulatory genes. Their products can be either activator or repressor. (2) The set of genes regulated by the same regulatory gene product is called a regulon. If a gene product regulates its own expression, it is said to be autoregulated. (3) Bacterial genes are often arranged in an operon which consists of a promoter region, an operator region and several structure genes. The mRNA of bacteria are made on a number of genes whose products perform related functions. This kind of mRNA is called polycistronic mRNA. Transcriptional regulation (4) There are two general types of transcriptional regulation : i. In negative regulation, a repressor binds to an operator and turns the operon off by preventing RNA polymerase from using or access the promoter. An operator sequence can be close to (up- or downstream), or even overlapping the promoter. ii. In positive regulation, an activator binds to the upstream of the promoter at an upstream activator site (UAS), where it can help RNA polymerase bind to the promoter or help open the promoter after the RNA polymerase binds. Two general types of transcriptional regulation Lac operon A. Bacteria respond to rapidly changing environments B. Examples: a. Lac operon(乳醣操縱組)of E. coli(大腸桿菌) 1. promoter sequence(起動子序列)– RNA polymerase (RNA聚合酶) 2. operator sequence(操縱子序列)– repressor protein 3. structural genes (構造基因)– Z (betagalactosidase; 半乳醣分解酶), Y (permease; 滲透酶) and A (transferase; 轉移酶) b. regulator gene(調控基因)– repressor protein (抑制蛋白質) The regulation of gene expression of Lac operon Operon Operator Regulatory gene Promoter Lactose-utilization genes DNA mRNA Protei repressr RNA polymerase cannot attach to promoter Active repressor OPERON TURNED OFF (lactose absent) DNA RNA polymerase bound to promoter mRNA Protein Repressor Lactose Inactive repressor Enzymes for lactose utilization OPERON TURNED ON (lactose inactivates repressor) The β- galactosidase reaction The lac control region Diauxic growth curve of E. coli grown with a mixture of glucose and lactose The interaction of promoters and CAP proteins in Lac operon A. CAP proteins are involved in positive regulation a. positive regulation – activator(活化子) b. CAP – catabolite activator proteins(降解物活化蛋白質) c. CAP binding site d. cAMP – cyclic adenosine monophosphate(環腺嘌呤二磷酸) e. CAP/cAMP complex – increasing the efficiency the ability of RNA polymerase binds to promoter. B. Catabolite repression(降解物抑制)– enabling E. coli to use glucose (葡萄糖) preferentially for energy even in the presence of lactose or other complex sugar. a. decreasing the level of cAMP b. permease - nonfunctional Positive Control of lac Operon • Positive control of lac operon by a substance sensing lack of glucose that responds by activating lac promoter – The concentration of nucleotide, cyclic-AMP, rises as the concentration of glucose drops The phosphoenolpyruvate (PEP)-dependent sugar phosphotransferase system (PTS) • Both HPr and IIA are the components of the PTS, which is responsible for transporting certain sugars, including glucose. Catabolite repression of the lac operon • Exogenous glucose inhibits both cAMP synthesis and the uptake of other sugars, such as lactose. • Components of the cascade : - HPr, the phosphotransferase (for histidine protein) transfers the phosphate from IIAGlc~P to sugar as the sugar is transported. - IIAGlc protein has two forms * IIAGlc~P activates adenylate cyclase to make cAMP. * IIAGlc inhibits sugarspecific permease that transport sugar Upstream activator site (UAS) 1. The αCTD (carboxyl terminus of the α subunits) binds to UP element (UAS), and αNTD binds to subunit. (A, B) 2. Some promoters lack a -35 sequence and instead have what is called extended -10 sequence. This sequence is recognized not byσ4 but, rather by σ3. (C) Hypothesis for CAP-cAMP activation of lac transcription Proposed CAP-cAMP Activation of lac Transcription • The CAP-cAMP dimer binds to its target site on the DNA • The aCTD (a-carboxy terminal domain) of polymerase interacts with a specific site on CAP(ARI: activation region I) • Binding is strengthened between promoter and polymerase • (The α–subunit N-terminal and C-terminal domains (αNTD and α–CTD, respectively) fold independently to form two domains that are tethered together by a flexible linker.) V. Regulation of gene expression 2. Posttranscriptional regulation – Gene expression can be regulated by (1) Inhibition of the translation of the gene even after mRNA is made (translational regulation). (2) Degradation of mRNA as soon as it is made or before it can be translated . (3) The protein product may be degraded by other protein, called protease. (4) By feedback inhibition – The final product inhibits enzyme activity of the first reaction in a pathway. V. Regulation of gene expression 3. Introns and inteins: (1) some genes have intervening sequence in the region of DNA encoding a RNA or protein. These sequence can move from one DNA to another. These sequences must be spliced out of RNAs and proteins after they are made to restore the function of RNAs or proteins. i. The intervening sequences that splice themselves out of RNA are called introns which are much more common in eukaryotic cells. ii. The intervening sequences that splice themselves out of protein are called intein. Feedback inhibition regulation VI. Expression vectors @ The cloning vectors designed to express (made) large amounts of proteins for biochemical or structural analysis. 1. Besides the elements of cloning vectors, expression vectors should have a promoter including operator, TIR including ATG, SD sequence and termination codon. 2. The gene or DNA sequence inserts into cloning site must be in-frame with ATG. 3. For easy purification of expressed protein, some affinity tags are also include in the vectors. (1) Histidine tag – DNA sequence encoding six histidine amino acids i. Histidines binds strongly to nickel, and so the protein contains histidines will bind to a column containing nickel. ii. Then the bound protein can be eluted by washing the column with high concentration of imidazole, which also binds to nickel and so will displace the Hist tag. (2) Other tag, such as glutathione S-transferase (GST) is used often. VI. Expression vectors Use pET-15b as an example. VI. Expression vectors Transcriptional and tranlational fusions to express lacZ VII. Some methods for studying gene expression - Northern blotting Buffer (20 X SSC) /1 L, pH 7.0 : 175.3 g of sodium chloride; 88.2 g 0f sodium citrate Northern Blots • You have cloned a cDNA – How actively is the corresponding gene expressed in different tissues? – Find out using a Northern Blot • Obtain RNA from different tissues • Run RNA on an denatureing agarose gel (usually containing formaldehyde) and blot to membrane • Hybridize to a labeled cDNA probe – Northern plot tells abundance of the transcript – Quantify using densitometer • Cytoplasmic mRNA isolated from 8 rat tissues probed with GPDH (glyceraldehyde-3-phosphate dehydrogenase) VII. Some methods for studying gene expression – Reverse transcription VII. Some methods for studying gene expression - Primer extension • Start with in vivo transcription, harvest cellular RNA containing desired transcript • Hybridize labeled oligonucleotide [18nt] (primer) • Reverse transcriptase extends the primer to the 5’-end of transcript • Denature the RNA-DNA hybrid and run the mix on a high-resolution DNA gel • Can estimate transcript concentration also VII. Some methods for studying gene expression - S1 nuclease mapping Use S1 nuclease mapping to locate the ends of RNAs and to determine the amount of a given RNA in cells at a given timeLabel a ssDNA probe that can only hybridize to transcript of interest - Probe must span the sequence start to finish - After hybridization, treat with S1 nuclease which degrades ssDNA and RNA - Transcript protects part of the probe from degradation - Size of protected area can be measured by gel electrophoresis * Amount of probe protected is proportional to concentration of transcript, so S1 mapping can be quantitative S1 Mapping the 5’ End Real-Time PCR 1. Real-time PCR quantifies the amplification of the DNA as it occurs 2. As DNA strands separate, forward and reverse primers anneal to DNA strand as that in regular PCR reaction. 3. A fluorescent-tagged oligonucleotide binds to part of one DNA strand Fluorescent Tags in Real-Time PCR 1. This fluorescent-tagged oligonucleotide serves as a reporter probe – Fluorescent tag at 5’-end – Fluorescence quenching tag at 3’-end 2. With PCR rounds, the 5’ tag is separated from the 3’ tag 3. Fluorescence increases with dNTPs incorporation into DNA product 4. The whole process takes place inside a fluorimeter that measure of the fluorescence of tag, which is in turn is a measure of the progress of the PCR reaction (in real time) 5. The reaction can be coupled to RTPCR VII. Some methods for studying gene expression –Biochip (Microarray ) Run-Off Transcription • DNA fragment containing gene to transcribe is cut with restriction enzyme in middle of transcription region • Transcribe the truncated fragment in vitro using labeled nucleotides, as polymerase reaches truncation it “runs off” the end • Measure length of run-off transcript compared to location of restriction site at 3’-end of truncated gene • Size of run-off transcript locates transcription start site • Amount of transcript reflects efficiency of transcription Nuclear Run-On Transcription • Isolate nuclei from cells, allow them to extend in vitro the transcripts already started in vivo in a technique called run-on transcription • RNA polymerase that has already initiated transcription will “run-on” or continue to elongate same RNA chains • Effective as initiation of new RNA chains in isolated nuclei does not generally occur, one can be fairly confident that any transcription observed in the isolated nuclei is simply a continuation of transcription that was already occurring in vivo • Therefore, the transcripts should reveal not only transcription rates but also give an idea about which genes are transcribed in vivo. VII. Some methods for studying gene expression – RNA interference (RNAi) 1. Also called cosuppression and posttrancriptional gene silencing (PTGS) 2. RNA interference occurs when a cell encounters dsRNA from a virus, a transposon, or a transgene (or experimentally added dsRNA). 3. This trigger dsRNA Is degraded into 21~23-nt fragments (siRNA) by an RNaseIII-like enzyme, Dicer. 4. The double-stranded siRNA, with Dicer and the associated protein R2D2, constitute a complex (complex B). 5. Complex B delivers the siRNA to the RISC loading complex (RLC), which probably separates the two strands of the siRNA and transfers the guide strand to the RNA-induced slicing complex (RISC), which includes a protein called Argonaute2 (Ago2). VII. Some methods for studying gene expression – RNA interference (RNAi) 6. The guide strand of the siRNA then base-pairs with the target mRNA in the active site in the PIWI domain of Ago2, which an RNase H-like enzyme also known as slicer. 7. Slicer cleaves the target mRNA in the middle of the region of its base-pairing with siRNA. 8. In an ATP-dependent step, the cleaved mRNA is ejected from the RISC, which can then accept a new molecule of mRNA to be degraded. RNA interference (RNAi) RNA interference (RNAi) shRNA (siRNA); hRluc (Renilla luciferase)