* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Document
Gene therapy of the human retina wikipedia , lookup
Genetically modified crops wikipedia , lookup
Transposable element wikipedia , lookup
Non-coding DNA wikipedia , lookup
Human genome wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Point mutation wikipedia , lookup
History of RNA biology wikipedia , lookup
Genetic engineering wikipedia , lookup
Genomic imprinting wikipedia , lookup
Short interspersed nuclear elements (SINEs) wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Polyadenylation wikipedia , lookup
Minimal genome wikipedia , lookup
Protein moonlighting wikipedia , lookup
Gene desert wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Genome (book) wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Gene nomenclature wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Messenger RNA wikipedia , lookup
RNA interference wikipedia , lookup
RNA silencing wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Microevolution wikipedia , lookup
Non-coding RNA wikipedia , lookup
Gene expression programming wikipedia , lookup
Helitron (biology) wikipedia , lookup
Genome evolution wikipedia , lookup
Designer baby wikipedia , lookup
Mir-92 microRNA precursor family wikipedia , lookup
Epigenetics of human development wikipedia , lookup
History of genetic engineering wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Epitranscriptome wikipedia , lookup
Gene expression profiling wikipedia , lookup
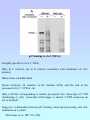
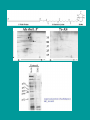
C4 dicots Amaranth Flaveria Ribulose 1,5-Bisphosphate Carboxylase (Rubisco) Reactions: CO2 + H2O RuBP (5C) 2 X 3-P-Glycerate (3C) + 2 H+ Rubisco O2 + H2O RuBP (5C) Rubisco 1 X 3-P-Glycerate (3C) + 2H+ + 1 X 2-P-Glycolate (2C) Located in chloroplasts: 6 Large Subunits (LSU), 55 kDa. rbcL gene Encoded on chloroplast genome Contains substrate (CO2 and O2) binding site. 6 Small Subunits (SSU), 12-18 kDa. RbcS gene Encoded in nuclear genome as gene family Synthesized as precursor, 20 Kda, with plastid transit sequence Transported to chloroplasts Possibly for regulation and assembly How are rbcL and RbcS genes encoding Rubisco regulated in C4 plants? Regulated by leaf development? Regulated by light? Transcriptional? Post-transcriptional? A B C A B In vivo protein synthesis and mRNA levels Polysome analysis: RbcS and rbcL mRNAs are associated with polysomes in L and in D+5hr L plants RbcS and rbcL mRNAs are not associated with polysomes in D plants Regulation in response to light occurs at the level of translation initiation Rubisco rbcL and RbcS genes are regulated post-transcriptionally in C4 plants Rubisco gene expression in C4 leaves is influenced by: Light Leaf development Photosynthesis Genes encoding other C4 enzymes show independent patterns of regulation What proteins regulate translation or stability of the Rubisco mRNAs, and what cis-acting sequences are recognized by these proteins? Two approaches: Biochemical analysis (search for mRNA binding proteins) Transgenic C4 plants (Search for cis-acting regulatory sequences) p47 binding to rbcL 5'RNA: Is highly specific to rbcL 5’ RNA Only in L extracts, not in E extracts (correlates with translation of rbcL protein) Observed as a doublet band Occurs between -14 (relative to the initiator AUG ) and the end of the processed rbcL 5’ UTR at –66 Only to RNAs corresponding to mature processed rbcL transcripts (5' UTR terminating at –66); transcripts with longer or shorter 5’UTR sequences do not to bind p47 Suggests a relationship between p47 binding, transcript processing, and rbcL translation in L-plants McCormac et al., JBC 276, 2001 pSK+ to generate transcripts for in vitro translation (T3) pBI221 for Biolistic (gene gun) transient expression (CaMV) pGA482 for expression in stably transformed C4 plants (CaMV) Graduate Students: Jingliang Wang Ping Xu Amy Swec Joe Boinski John Long Amy Corey Jianxin Wang Hanz Litz Vince Ramsperger Sabrina Picinic Francois Faure Postdocs: Minesh Patel Robert Givens Mei-Hui Lin Dennis McCormac Technicians: Sejal Patel Xiaowen Jang Collaborators: John Carr (Cambridge) Bill Taylor (Canberra) Li Ping Lin (Beijing) V. James Hernandez (UB) Derek Taylor (UB) Jeremy Bruenn (UB) Mary Bisson (UB) Funding: NSF Biochemistry of gene expression International programs USDA Photosynthesis Plant genetic mechanisms Undergraduates: Jackson Buss Elizabeth Herric Theresa Flood Francois Faure Kwan Lau Eric Rydzik Paul Nordone Marina Boruk Jeff Burkwitz Janet Lam Jay Friedman Melinda Bauman Renata Mazzei Diane Watkins Matt Bernhard Peter Depowski Curtis Erickson