* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Regulation of Gene Expression
Deoxyribozyme wikipedia , lookup
Genomic imprinting wikipedia , lookup
X-inactivation wikipedia , lookup
Gene nomenclature wikipedia , lookup
Biology and consumer behaviour wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Gene desert wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Epigenetics in learning and memory wikipedia , lookup
History of genetic engineering wikipedia , lookup
Non-coding DNA wikipedia , lookup
Minimal genome wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Polyadenylation wikipedia , lookup
Genome evolution wikipedia , lookup
Genome (book) wikipedia , lookup
Gene expression programming wikipedia , lookup
Ridge (biology) wikipedia , lookup
Messenger RNA wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
RNA interference wikipedia , lookup
Transcription factor wikipedia , lookup
History of RNA biology wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Point mutation wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Microevolution wikipedia , lookup
Short interspersed nuclear elements (SINEs) wikipedia , lookup
RNA silencing wikipedia , lookup
Designer baby wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Mir-92 microRNA precursor family wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Gene expression profiling wikipedia , lookup
Epitranscriptome wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Non-coding RNA wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
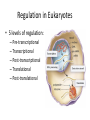
SBI4U1 Regulation of Gene Expression Gene Regulation: control of the level of gene expression -whether a gene is active or inactive -determines the level of activity and the amount of protein that is available. Constitutive Genes: genes that are always active and expressed at constant levels “ housekeeping genes” Regulation of gene expression in prokaryotes occurs at three different levels • Transcription * most common during initiation • Translation • After protein has been synthesized Recall? Promoter region is where RNA polymerase binds to DNA to initiate trancription What is an Operon? A cluster of genes under the control of 1 promoter -these genes are all transcribed together Let’s use E. Coli as an example…they use different sugars as a source of energy The “Lac Operon” • Genes that encode the enzymes needed to break down the sugar lactose • Composed of 2 regions: regulatory region & coding region Enzymes which break down lactose Promoter Operator Z Y A Controls whether gene in transcribed or not RNA polymerase binds here to start making mRNA copy Here’s how it works… • In the absence of lactose, the lac repressor protein binds to the operator, preventing RNA polymerase from binding to the promoter to initiate transcription. • In the presence of lactose, an activator binds to the repressor so it can no longer bind to the operator. Transcription of the genes can therefore occur. The “Trp Operon” • If tryptophan is present; no transcription Genes synthesize tryptophan. The operon is transcribed until sufficient tryptophan is present. Promoter Operator E D C Trp repressor binds to promoter and inhibits transcription B A “Trp Operon” contd.. The trp (tryptophan) operon, however, is normally active until a repressor turns it off. It contains: • a coding region with five genes for enzymes required for tryptophan synthesis • a regulatory region with a promoter and an operator The trp operon contains five genes that are involved in the synthesis of tryptophan. This operon is normally transcribed until the cell has sufficient tryptophan. Then the trp repressor binds to the promoter and inhibits transcription. Regulation in Eukaryotes • 5 levels of regulation: – Pre-transcriptional – Transcriptional – Post-transcriptional – Translational – Post-translational Pre-Transcriptional/Transcriptional • DNA in highly condensed chromatin is not transcribed – Chromatin is physical barrier to proteins needed to synthesize pre-mRNA • Can be loosened so transcription can occur via chromatin remodelling complexes • Transcription factors are a set of proteins needed to RNA polymerase to bind to promoter – Activators bind to transcription factors, RNA polymerase, enhancers Post-Transcriptional/Translation • mRNA level • 5’ cap/poly-A tail may not be added – mRNA does not leave nucleus – Or is degraded in cell • Small RNA molecules control gene expression via RNA interference – Micro RNA (miRNA) and small interfering RNA (siRNA) turn off gene expression Post-Translational • Polypeptide level • They must be activated by modifications • E.g. Insulin is folded into 3D structure – Removal of amino acids activates it – Leaves two polypeptide chains • Regulating length of protein in cell also regulates the gene – Adding a chain of – molecules can signal protein degradation Learning Expectations... • What is gene regulation? • Where does it occur in a cell? • How do the Lac and Trp operons work? – Compare/contrast • Difference in gene regulation in prokaryotes and eukaryotes