* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Protein degradation and regulation
Genetic code wikipedia , lookup
Evolution of metal ions in biological systems wikipedia , lookup
Transcriptional regulation wikipedia , lookup
Ancestral sequence reconstruction wikipedia , lookup
Gene regulatory network wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Point mutation wikipedia , lookup
Ribosomally synthesized and post-translationally modified peptides wikipedia , lookup
Ultrasensitivity wikipedia , lookup
Biosynthesis wikipedia , lookup
Metalloprotein wikipedia , lookup
Mitogen-activated protein kinase wikipedia , lookup
Expression vector wikipedia , lookup
Magnesium transporter wikipedia , lookup
Gene expression wikipedia , lookup
Amino acid synthesis wikipedia , lookup
Interactome wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Acetylation wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Biochemical cascade wikipedia , lookup
Biochemistry wikipedia , lookup
Protein purification wikipedia , lookup
Protein structure prediction wikipedia , lookup
Signal transduction wikipedia , lookup
Paracrine signalling wikipedia , lookup
Western blot wikipedia , lookup
Protein–protein interaction wikipedia , lookup
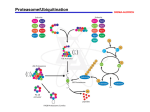
Protein Degradation and Regulation Ubiquitin/Proteasome Pathway Guo Peng, Luo Tong and Yang Kong 2002.12.16 I. Introduction This pathway is the major non-lysosomal process responsible for the breakdown of most short and longlived proteins in mammalian cells. For example, in skeletal muscle, the system is responsible for the breakdown of the major contractile proteins, actin and myosins. In addition, the pathway also controls various major biological events: cellcycle progression, oncogenesis, transcriptional control, development and differentiation, signal transduction, receptor downregulation and antigen processing, via the breakdown of specific proteins. two main steps in the pathway 1. 2. covalent attachment of a polyubiquitin chain to the substrate; specific recognition of this signal, and degradation of the tagged protein by the 26S proteasome. Cellular functions of protein degradation The elimination of damaged proteins: environmental toxins, translation errors and genetic mutations can damage proteins. Misfolded proteins are highly deleterious to the cell because they can form non-physiological interactions with other proteins. Repair proteins called chaperones can, in many instances, restore the native conformation of misfolded proteins. However, if a damaged protein is not repaired, it is degraded in specialized organelles such as the ysosome, and by the Mislocalized proteins and stoichiometric excess Some proteins are stabilized only when they are bound to their natural partners. This ensures that they are present only at stoichiometric levels. Consequently, the overexpression of specific ribosomal proteins can lead to degradation because of their failure to assemble into the ribosome. Similarly, proteinsthat are mislocalized may be degraded because they are unable to form interactions that normally stabilize Retro-translocation Proteins that enter the secretory pathway and fold improperly in the endoplasmic reticulum are transported back to the cytosol where they are recognized and degraded by the ubiquitin/proteasome pathway. Degradation of foreign proteins The immune system is a surveillance mechanism that can recognize foreign proteins and degrade them. An essential feature of this system is the ability to distinguish ‘self’ from ‘non-self’. The MHC class I antigen presenting cells display peptide fragments that are derived from the foreign protein, to cytotoxic T cells. The generation of these peptides Degradation of regulators: Many regulators of cell growth and development are highly unstable proteins, whose stability is controlled by the ubiquitin/proteasome pathway. Substrates of this pathway include p53, Rb, cyclins, CDK inhibitors, transcription factors, and signaltransducing molecules. Distinct targeting complexes accomplish the The generation of active proteins Enzymes whose activities can be deleterious to the cell are often expressed as precursors that are catalytically inactive. The proteolytic cleavage of the precursor generates an active enzyme. For instance, proteases that are present in the digestive tract, and those that function in the lysosome, are initially synthesized as precursors. Ubiquitin, and catalytic subunits of the proteasome are also expressed as precursors that are proteolytically processed to yield catalytically active subunits. The recycling of amino acids: Proteases are required for the generation of free amino acids from short peptides that are generated by the proteasome and other intracellular proteases. In many microorganisms dipeptidases and other proteases that hydrolyze short amino acid chains are secreted to generate free amino acids that can be readily imported into the cell.The availability of free amino acids and di-peptides can allosterically regulate the activity of a specific E3 protein, which in turn controls the levels of a transcription factor that is required for inducing amino acid biosynthetic pathway genes. II. Protein Degradation Ubiquitin Ubiquitin is a highly conserved protein (3 aa exchanges from yeast to men) Ubiquitin is composed of 76 aa Attachment site to target protein on ubiquitin is C-terminus Bond is formed to side chain of Lys of target protein Attachment is performed by array of enzymes (E1, E2, E3, E4) Subsequently, poly-ubiquitin chains form via binding of further molecules to Lys side chains (Lys48 > 6, 11, 29, 63) of primary ubiquitin Enzymes of the Ubiquitination E1: • ubiquitin-activating enzyme. • exists as two isoforms of 110- and 117-kDa, which derive from a single gene and are found in both the nucleus and cytosol. Inactivation of this gene is lethal. • In mammals there is a single E1. E2: • Ubiquitin-conjugating enzymes. • E2s are a superfamily of related proteins. There are eleven E2s in yeast, and 20-30 E2s in mammals. E3s: • Ubiquitin-protein ligases. • E3s play a key role in the ubiquitin pathway, as they are responsible for the selective recognition of protein substrates. • E3 ligases can be subdivided into at least six subtypes. E4: • catalyzes the efficient polymerization of very long polyubiquitin chains, it has been characterized in yeast. How is ubiquitin activated? C-terminus of ubiquitin gets adenylated Rearrangement to intermolecular thioester with a E1 (activation enzyme) Transfer of activierted ubiquitin from E1 to E2 (ubiquitin-conjugating enzyme) (thioester bond) Transfer form E2 via E3(ubiquitin ligase) to target enzyme Process of ubiquitin activated Combinatorial nature of ubiquitination Modes of recognition of protein substrates by the different E3s Which signals lead to ubiquitination? Genetic program (amino acids) • N—end rule N—terminal amino acid: D,R,L,K,F (< minutes); A,G,M,S,V (>10 hours) • Sequence of significant hydrophobicity(疏水性) • “PEST” sequences (sequences rich in Pro, Asp, Glu, Ser and Thr) Phosphorylation of Ser and Thr Binding to adaptor proteins(衔接蛋白) Protein damage • Processing • Oxidation of Cys and Met • Age-dependent modifications of side chains: hydrolsis(水解), deaminations(脱氨基), racemizations(外消旋化), disulfide bond breaks(二硫键簖裂), ketoamines(氯氨酮) … • Wrong folding Themes and Variations on Ubiquitylation Pay attention Ubiquitination is an important and widespread post-translational modification of proteins, which resembles phosphorylation. Very importantly, ubiquitination is not only a degradation signal, but also directs proteins to a variety of fates which include roles in ribosomal function, in DNA repair, in protein translocation, and in modulation of structure or activity of the target proteins. In order to be efficiently degraded, the substrate must be bound to a polyubiquitin degradation signal that comprises at least four ubiquitin moieties, These signals are usually determined by short regions in the primary sequence of the targeted protein. The nature of the N-terminal amino acid of a protein (N-end rule) may determine its rate of polyubiquitination and subsequent degradation. Monoubiquitination and multimubiquitination Deubiquitination enzymes Eukaryotic cells also contain DUBs (DeUBiquitinating enzymes), which are encoded by the UCH (Ubiquitin Carboxyl-terminal Hydrolases) and the UBP (UBiquitin-specific Processing proteases) gene families. UCHs are relatively small proteins (< 40-kDa);in contrast, UBPs are 50-250-kDa 8proteins and constitute a large family. Genome sequencing projects have identified more than 90 DUBs . Possible roles for DUB enzymes Editing proofread Disassembly Recycling Processing Basic features of proteasome Essential and ubiquitous intracellular protease Degrades most of cytoplasmatic, nuclear and membrane , nuclear and membrane proteins (> 90 %) Virtually all target proteins are marked by ubiquitin first Ubiquitin is recycled, not cleaved Central processes with proteasome involvement are mitosis, antigen presentation, activation and degradation of transcription factors and regulation of developmental processes. Eukaryotic proteasomes are large protein complexes of ~ 2000 kDa, consisting of a “core” and a “cap” region Prokaryotes lack ubiquitin system and possess no cap region Schematic representation of the eukaryotic Core particle is composed of four 7-membered rings. Two types of subunits (25 kDa): αand β, all differ . Subunits are similar in structure, different in sequence. only only β subunits are catalytically active . Cap region regulates activity, performes the energy dependent steps. The structure of proteasome Processing via the proteasome •Length of produced peptides: 3-23 amino acids •Average length of peptides: 7-9 amino acids •Peptide composition of given protein stays constant •Protein is completely degraded before import of next protein •Peptides produced by proteasome are further degraded by other roteases and aminopeptidases (Tricorn, Multicorn, Thimet, TPPII) • Proteasome and immune system function: •Peptides of 8-9 amino acids in length are transported to the cell surface via the ER presented on the cell surface via MHC class I – molecules Central position of the proteasome Site of intracellular degradation Ubiquitin—mediated degradation of cytosolic and membrane proteins occurs in the cytosol and on the cytosolic face of the ER membranes. Although components of the system have been localized to the nucleus, conjugation and degradation have not been demonstrated in this organelle. Alternative pathways The 26S proteasome is not an absolute ubiquitin-dependent proteolytic enzyme, as it also degrades nonubiquitinated substrates. protein c-Fos lysosomal ODC ubiquitination c-Jun proteasome calpai III. Protein Regulation (I). General regulation Alternation of E1, E2s and proteasome in their activity will affect many substrates. One is the up-regulation of the ubiquitin pathway to achieve bulk degradation of skeletal muscle proteins that occurs in different pathophysiological conditions such as fasting(禁 食), cancer cachexia(癌血症), severe sepsis(脓毒), metabolic acidosis(代谢酸中毒)。 The second example of a change in the general components of the system occurs following treatment with IFN-r. This cytokine induces changes in the subunit composition of the 20S proteasomal complex. Consequently, the antigenic peptides that are generated following proteosomal degradation have higher affinity for the presenting MHC class I molecules and for the cytotoxic T-cell receptor . (II). Specific regulation A. Regulation by modification of the substrate: Phosphorylation of many substrates is required for their recognition by their E3s. Conversley, similar modification of many other proteins prevents this. Substrates that require prior phosphorylation include the yeast G1 cyclins(细胞周期蛋白), Cln2 and Cln3, the yeast cyclin—dependent kinase (CDK) inhibitors, Sic1 and Far1. Degradation of the proto-oncogene c-mos by the ubiquitin pathway is inhibited by phosphorylation on Ser. Interestingly, activation of c-mos leads to phosphorylation and stabilization of c-fos, another substrate of the ubiquitin pathway. B. Regulation by modulation of ubiquitination activity: Regulated degradation of specific classes of substrates could be achieved by modulation of the activity of the ubiquitination machinery. For example, it has been shown recently that degradation of mitotic regulators by the APC(抗原 呈递细胞) is regulated by different activators and inhibitors and by phosphorylation C. Regulation by ancillary proteins: Several viral proteins exploit the ubiquitin system by targeting for degradation cellular substrates which may interfere with propagation of the virus. In some instances, the viral protein functions as a bridging‘ element between the E3 and the substrate, thus conferring recognition in trans. The prototype of such a protein is the high risk HPV oncoprotein(人乳头癌蛋白)E6 which interacts with an E6AP HECT domain E3, and with the tumor suppressor protein p53. This interaction targets p53 for rapid degradation and, thus, most probably prevents stress signalinduced apoptosis and ensures further replication propagation of the virus . In a different case, the Vpu protein of the HIV-1 virus is recognized by the F-box protein, b-TrCP. Vpu also binds to the CD4 receptor in the ER of Tcells infected by the virus. This leads to ubiquitination and subsequent degradation of CD4 by the SCFb-TrCP complex, thus enabling the virus to escape from immune surveillance. D. Regulation by masking of a degradation signal: The presence of either one of two transcription factors, MATa1 and MATa2, determines the mating type of haploid yeast cells. The diploid cell expresses both a1 and a2 that form a heterodimer with distinct DNA-binding specificity. In haploid cells, the two factors are rapidly degraded by the ubiquitin system. Degradation of a2 requires two degradation signals, Deg1 and Deg2. Strikingly, both a1 and a2 are stabilized by heterodimerization.For a2 at least, it has been shown that residues required for interaction with a1 overlap with the Deg1 degradation signal and it is possible that binding of a1interferes with the degradation of a2 by masking the ubiquitin recognition signal. IV. Conclusions and future perspectives Only a few targeting signals have been identified, and the mechanisms that underlie the regulation of the system are still largely unknown? While the system has been implicated in the pathogenesis of several diseases, the underlying mechanisms, as well as its potential involvement in many other diseases, are still an enigma? Why are there so many ubiquitinating enzymes if prior modifications such as phosphorylation or damage are triggering events? Do DUBs show substrate specificity, perhaps by regulating the levels of ubiquitination of specific subsets of proteins? What are the binding sites for polyubiquitin chains on the microtubules and on the proteasome itself? What is the role of K29-and/or K63-linked polyubiquitin chains in the cell? THANK YOU !