* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Post-translation
Biochemical switches in the cell cycle wikipedia , lookup
Cytokinesis wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Endomembrane system wikipedia , lookup
Phosphorylation wikipedia , lookup
Magnesium transporter wikipedia , lookup
Signal transduction wikipedia , lookup
Histone acetylation and deacetylation wikipedia , lookup
Protein folding wikipedia , lookup
Protein (nutrient) wikipedia , lookup
Protein structure prediction wikipedia , lookup
Protein phosphorylation wikipedia , lookup
Intrinsically disordered proteins wikipedia , lookup
Protein moonlighting wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Protein–protein interaction wikipedia , lookup
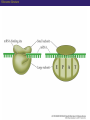
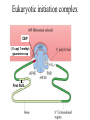
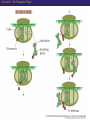
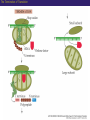
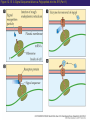
Proteomics and posttranslational modifications Xiaozhong Peng Department of Molecular Biology and Biochemistry National Laboratory of Medical Molecular Biology CAMS & PUMC Protein Translation Ribosome Structure The Initiation of Translation Preinitiation complex Initiation complex Mechanism of translation initiation. Eukaryotic initiation complex CBP ( 5’ cap) 7-methylguanosine cap First AUG Translation: The Elongation Stage The Termination of Translation Figure 12.13 A Polysome (Part 1) Figure 12.13 A Polysome (Part 2) Figure 12.14 Destinations for Newly Translated Polypeptides in a Eukaryotic Cell Pre-translation: take place at the level of amino acyl-tRNA prior to polymerization. Co-translation: take place during polymerization. Post-translation: take place after the completed protein has been released from the polysome. Post-translational modifications Post-translational modifications ● N-terminal or C-terminal modification – Removal of N-formylmethionine – N-acetylation (50% of eucaryotic proteins) ● N-terminal and C-terminal processing – Maturation, proteolytic processing ● Modification of individual amino acids Figure 12.16 Posttranslational Modifications to Proteins – Phosphorylation – Glycosylation – Methylation – Farnesylation ● Protein splicin: Intein ● More than 200 known posttranslational Modifications have been reported. --Gudepu, R.G.& Wold,F.(1998)in Proteins:Analysis and Design,ed.Angeletti, R.H.(Academic, San Diego),pp.121-207. ● More than 300 different types of PTMs are currently known and new ones are regularly discovered. --Ole Norregaard Jensen. Current Opinion in Chemical Biology 2004,8:33-41. ● Protein Ubiquitination The Nobel Prize in Chemistry 2004 "for the discovery of ubiquitin-mediated protein degradation" Aaron Ciechanover Avram Hershko Technion – Israel Institute of Technology Haifa, Israel Technion – Israel Institute of Technology Haifa, Israel Irwin Rose University of California Irvine, CA, USA Proteins build up all living things: plants, animals and therefore us humans. In the past few decades biochemistry has come a long way towards explaining how the cell produces all its various proteins(at least five Nobel Prizes have been awarded in this area). But as to the breaking down of proteins, not so many researchers were interested.Aaron Ciechanover,Avram Hershko and Irwin Rose went against the stream and at the beginning of the 1980s discovered one of the cell's most important cyclical processes, regulated protein degradation. For this, they are being rewarded with this year's Nobel Prize in Chemistry. This year's Nobel Laureates in chemistry, Aaron Ciechanover, Avram Hershko and Irwin Rose, have contributed ground-breaking chemical knowledge of how the cell can regulate the presence of a certain protein by marking unwanted proteins with a label consisting of the polypeptide ubiquitin. Proteins so labelled are then broken down – degraded – rapidly in cellular "waste disposers" called proteasomes. Protein Degradation: Schoenheimer: a pioneer in this field! 1942--isotope tracer techniques—indicated that proteins in animals are continuously synthesized and degraded and therefore are in a Dynamic state. Degradation needs no energy-or does it? It doesn’t: Trypsin: a type of cell organelle: Lysosome It does: 1.Simpson, 1953: release of amino acids from cultured liver slices was energy-dependent. 2.Hershko and Tomkin, 1971: energy-dependent degradation of the Enzyme tyrosine aminotransferase in cultured hepatoma cells. 3.Ciechanover, 1997: tyrosine aminotransferase degradation is indeed ubiquitin-mediated. The Label is ubiquitin: ● Was first isolated from bovine thymus (calf sweetbread) by Goldstein in 1975. ● Busch found “protein A24”-histone H2A+ubiquitin.??? ● Hunt and Dayhoff found in 1977 .Named from Latin ubique, “everywhere”. ● 76 amino acids peptide.found in numerous different tissues and organisms-but not in bacteria. Fig 1. Ubiquitin - a common polypeptide that represents the "kiss of death". The discovery of ubiquitin-mediated protein degradation: ● A major part of the work was done during a series of sabbatical leaves when Hershko and Ciechanover worked in Rose’s laboratory at the Fox Chase Cancer Center in Philadephia. Two surprising discoveries: ●in 1978, when Reticulocyte lysate system was passed over a DEAE cellulose column to remove the hemmoglobin, two fractions one contains APF-1(active principle of fraction1)-ubiquitin. ●in 1979,the second fractions subdivided by salt precipitation into two :one contains 450kDa protein-proteasome, and another contains E1-E3 enzymes. The Breakthrough in 1980: 125I-Labeled APF-1 125I-Labeled lysozyme, a-lactalbumin and globin Two novel enzymatic activities: 1. The E1 enzyme activates the ubiquitin molecule. This reaction requires energy in the form of ATP. 2. The ubiquitin molecule is transferred to a different enzyme, E2. 3. The E3 enzyme can recognise the protein target which is to be destroyed. The E2-ubiquitin complex binds so near to the protein target that the actual ubiquitin label can be transferred from E2 to the target. 4. The E3 enzyme now releases the ubiquitin-labelled protein. 5. This last step is repeated until the protein has a short chain of ubiquitin molecules attached to itself. 6. This ubiquitin chain is recognised in the opening of the proteasome. The ubiquitin label is disconnected and the protein is admitted and chopped into small pieces. Fig 2. Ubiquitin-mediated protein degradation Multi-step ubiquitin-tagging hypothesis: The Proteasome-the cell’s waste disposer ●A human cell contains about 30,000 proteasomes, can break down practically all proteins to 7-9-aa-long peptides. Fig 3. The cell's waste disposer, the proteasome. The black spots indicate active, protein-degrading surfaces. More recent research: ● Regulation of the cell cycle( for example) APC: anaphase-promoting complex To identify the E3 ligase responsible for H2A ubiquitination , They Developed an assay: in the presence of E1,E2,ATP and Flag-Ubiquitin(FUb), Hela nuclear protein fractions were tested for E3 Ligase activity with core histone or nucleosomal histone substrates. Chromatin Immunoprecipitation DNA-Protein Interaction Immunoprecipitation Histone Modification PCR Identification The schematic principle of ChIP Summary: ● Purified an E3 ubiquitin ligase complex that is specific for histone H2A. ● The complex, termed hPRC1L(human Polycomb repressive complex1-like), is composed of several Polycomb-group proteins including Ring1, Ring2, Bmi1and HPH2. ● hPRC1L monoubiquitinates nucleosomal histone H2A at lysine 119. ● Reducing the expression of Ring2 results in a dramatic decrease in the level of ubiquitinated H2A in HeLa cells. ● Chromatin immunoprecipitation analysis demonstrated colocalization of dRing with ubiquitinated H2A at the PRE and promoter regions of the Drosophila Ubx gene in wing imaginal discs. ● Removal of dRing in SL2 tissue culture cells by RNA interference resulted in loss of H2A ubiquitination concomitant with derepression of Ubx. Thus, their studies identify the H2A ubiquitin ligase, and link H2A ubiquitination to Polycomb silencing. Ubiquitin like molecule: ● Protein Glycosylations Figure 12.14 Destinations for Newly Translated Polypeptides in a Eukaryotic Cell Figure 12.15 A Signal Sequence Moves a Polypeptide into the ER (Part 1) Figure 12.15 A Signal Sequence Moves a Polypeptide into the ER (Part 2)