* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Introduction
Protein design wikipedia , lookup
Green fluorescent protein wikipedia , lookup
Homology modeling wikipedia , lookup
Protein domain wikipedia , lookup
Protein structure prediction wikipedia , lookup
Protein folding wikipedia , lookup
Protein moonlighting wikipedia , lookup
Bimolecular fluorescence complementation wikipedia , lookup
Protein mass spectrometry wikipedia , lookup
Intrinsically disordered proteins wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Protein purification wikipedia , lookup
Western blot wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
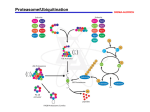
Introduction Turnover of cellular proteins was discovered in the 1930s in studies of Rudolf Schoenheimer, but it was in the 1960s that is became apparent that this was not just turnover, but a highly selective process. By the end of the 1970s two independent groups were working on two different topics: in the lab of Avram Hershko in Haifa, Israel, Hersko and Ciechanover were working on the ATP dependent degradation of the tyrosine aminotransferase. They isolated a protein and named it ATP-dependent proteolysis factor 1 (APF-1). On the other side of the Atlantic Ocean, Alexander Varshavsky, settled down in Boston and became interested in a DNA binding protein containing one C-terminus and interestingly two N-termini! The DNA binding protein was histone 2a and the other protein was identified as ubiquitin, a 76-residue ubiquitously (hence its name) expressed protein of unknown function that was described (as a free protein) by Gideon Goldstein and colleagues in 1975. In 1980, Keith Wilkinson, Michael Urban and Arthur Haas showed that APF-1 and ubiquitin were the same protein. Ubiquitin-Proteasome System (UPS) Ubiquitin conjugation to substrate proteins is one of the best-known post-translational modifications in the cell. It can not only destine its target proteins for degradation, but is also involved in gene transcription, endocytosis and DNA damage repair. The most well-known function is of course protein breakdown. The addition of four or more ubiquitins to a substrate target it to the proteasome (see figure). This mega-Dalton complex is responsible for unfolding and degrading proteins back to aminoacids. Ubiquitination is a consequence of ubiquitin transfer between different proteins: ubiquitin is first activated in an ATP dependent manner by an ubiquitin-activating enzyme, called E1 (see figure). Activated ubiquitin is then transferred via a thioester intermediate to an ubiquitin-conjugating enzyme, called E2. This activated E2 then acts in concert with an ubiquitin-ligase, called E3, to transfer the ubiquitin to a target substrate, forming an isopeptide bond between the ε-amino group of the Lysine residue of the substrate and the C-terminal Glycine residue of ubiquitin. To study the behaviour of ubiquitin in living cells, we fused the Green Fluorescent Protein (GFP) to the N-terminus of ubiquitin as was described before by Yewdell and Dantuma. In this system we are able to compare the ubiquitin and the proteasome in stably transfected living cells, because we already had a β1i-GFP (LMP2-GFP) cell line with a tagged proteasome subunit. Approach As mentioned above, we coupled GFP to the Nterminus of ubiquitin and generated stable cell lines expressing this fusion protein. We have done several tests to ensure that the construct does what it is supposed to do (binding covalently to target proteins) and does not do what it is not supposed to do (changing MHC class I expression or the cell cycle). We have tried to gain more insight in ubiquitin localization and mobility with the help of the fluorescence techniques Fluorescence Recovery After Photobleaching (FRAP) and Fluorescence Loss In Photobleaching (FLIP). For very fast FRAP measurements we have used a confocal equipped with an external bleaching laser to avoid changing mirrors, laser intensities etc. Diffusion and localization of the proteasome is not affected by proteasome inhibition, and FLIP experiments show that no exchange between the nucleus and the cytoplasm is possible (see example trace). Reference T. Groothuis and E. Reits, Manuscript submitted Fatal error Problems in the ubiquitin-proteasome system are thought to be involved in several diseases, mainly aggregation diseases like Huntington's, Alzheimer and Parkinson's disease. Extended CAG repeats in the involved proteins aggregate in the cytoplasm and the nucleus, and cannot be degraded by the proteasome although both ubiquitin and proteasome are present in the aggregates. Aggregates are formed by the cell to better cope with the misfolded proteins. On the horizon Therapies targeted at the proteasome may also help to treat cancers. Currently, potential therapeutic applications associated with inhibiting this enzyme are evaluated in clinical trials in a variety of solid and hematologic malignancies. Millennium is currently developing VELCADE™ (bortezomib) for injection, the first proteasome inhibitor to be studied in human clinical trials for any disease. The use of of this drug to treat refractory multiple myeloma has been granted fast-track status by the U.S. Food and Drug Administration (FDA), and it has also been granted Orphan Product Designation for the treatment of multiple myeloma. Fast Recovery Fluorescence Recovery After Photobleaching , or FRAP, is a non-invasive technique, which uses photobleaching (also termed fading). It occurs when a fluorophore permanently loses the ability to fluoresce due to photon-induced chemical damage and covalent modification of the fluorophore. Diffusing proteins in a living cell are partially bleached, after which recovery of fluorescence in the bleached spot is recorded. Read more about FRAP and its applications on the following page: More on FRAP >> Technicalities Molecular cloning techniques were used to construct the fluorescently labeled ubiquitin chimera Westernblot analysis and confocal microcopy confirmed expression of the recombinant protein FRAP with confocal microscopy was applied to investigate the localization and mobility of the proteasome and ubiquitin Metabolic labelling with radioactive 35-S Methionine/Cystein for protein turnover Electron microscopy generated ultrastructural insight