* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Abstract I. DLC1 encodes a RhoA GTPase
Survey
Document related concepts
Protein design wikipedia , lookup
Homology modeling wikipedia , lookup
Protein folding wikipedia , lookup
Protein structure prediction wikipedia , lookup
Western blot wikipedia , lookup
Degradomics wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Bimolecular fluorescence complementation wikipedia , lookup
Protein purification wikipedia , lookup
List of types of proteins wikipedia , lookup
Protein mass spectrometry wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Transcript
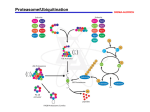
Abstract I. DLC1 encodes a RhoA GTPase-activating protein and tumor suppressor lost in cancer by genomic deletion or epigenetic silencing and loss of DLC1 gene transcription. We unexpectedly identified non-small cell lung cancer (NSCLC) cell lines and tumor tissue that expressed DLC1 mRNA yet lacked DLC1 protein expression. We determined that DLC1 was ubiquitinated and degraded by cullin 4A-RING ubiquitin ligase (CRL4A) complex interaction with DDB1 and the FBXW5 substrate receptor. siRNA-mediated suppression of cullin 4A, DDB1, or FBXW5 expression restored DLC1 protein expression in NSCLC cell lines. FBXW5 suppression-induced DLC1 reexpression was associated with a reduction in the levels of activated RhoA-GTP and in RhoA effector signaling. Finally, FBXW5 suppression caused a DLC1-dependent decrease in NSCLC anchorage-dependent and -independent proliferation. In summary, we identify a posttranslational mechanism for loss of DLC1 and a linkage between CRL4A-FBXW5-associated oncogenesis and regulation of RhoA signaling. II. Defining the full complement of substrates for each ubiquitin ligase remains an important challenge. Improvements in mass spectrometry instrumentation, computation and protein biochemistry methods have resulted in several new methods for ubiquitin ligase substrate identification. Here we used the parallel adapter capture (PAC) proteomics approach to study βTrCP2/FBXW11, a substrate adaptor for the SKP1-CUL1-F-box (SCF) E3 ubiquitin ligase complex. The processivity of the ubiquitylation reaction necessitates transient physical interactions between FBXW11 and its substrates, thus making biochemical purification of FBXW11-bound substrates difficult. Using the PAC-based approach, we inhibited the proteasome to 'trap' ubiquitylated substrates on the SCFFBXW11 E3 complex. Comparative mass spectrometry analysis of immunopurified FBXW11 protein complexes before and after proteasome inhibition revealed 21 known and 23 putatively novel substrates. In focused studies, we found that SCFFBXW11 bound, polyubiquitylated and destabilized RAPGEF2, a guanine nucleotide exchange factor that activates the small GTPase RAP1. High RAPGEF2 protein levels promoted cell-cell fusion and consequently multinucleation. Surprisingly, this occurred independently of the GEF catalytic activity and of RAP1. Our data establish new functions for RAPGEF2 that may contribute to aneuploidy in cancer. More broadly, this study supports the continued use of substrate trapping proteomics to comprehensively define targets for E3 ubiquitin ligases.