* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Chap 3
Gene regulatory network wikipedia , lookup
Community fingerprinting wikipedia , lookup
Bottromycin wikipedia , lookup
Polyadenylation wikipedia , lookup
Molecular cloning wikipedia , lookup
Transcription factor wikipedia , lookup
List of types of proteins wikipedia , lookup
Endogenous retrovirus wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Non-coding RNA wikipedia , lookup
Biochemistry wikipedia , lookup
Molecular evolution wikipedia , lookup
Non-coding DNA wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Expanded genetic code wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Messenger RNA wikipedia , lookup
Deoxyribozyme wikipedia , lookup
RNA polymerase II holoenzyme wikipedia , lookup
Eukaryotic transcription wikipedia , lookup
Promoter (genetics) wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Gene expression wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Genetic code wikipedia , lookup
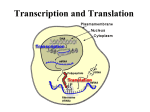
1 Chap. 3 The Nature of Living Things: How They Function 1. 2. 3. 4. 5. Purpose and expected outcomes The central dogma of molecular biology. The genetic code. How the genetic code is translated into proteins. How organisms metabolize food. The opportunities in metabolic pathways for genetic manipulation of organisms. Genetic bases of function 1. The function of an organism is an interplay of genetics and the environment. 2. Genes are expressed as proteins. 3. More than one protein can be expressed from one gene. What is the cent ral dogma? 1. The central dogma of molecular biology is the concept that information flow progresses from DNA to RNA to protein but not the reverse (Fig. 3-1) (“old” dogma). 2. Watson and Crick (who discovered the physical structure of DNA) first stated this dogma. 3. The technique of reversing the central dogma is the synthesis of a complementary DNA (cDNA) using an enzyme called “reverse transcript” (“new” dogma). 1. 2. 3. 4. 5. 6. 7. 8. DNA replication Replication must be executed to a high degree of fidelity, with errors occur at a low rate of 1 in 109 to 1 in 1010 base pairs. The first step to duplicate a double-stranded DNA molecule: the double helix unwinds so that each strand serves as a template for the synthesis of its complement. Each replicated DNA molecule would comprise one new strand and one parent strand (semiconservative). DNA replication starts at an “origin of replication (ori) ”. The unit or length of DNA that is replicated following one initiation event at one origin of replication is called a “replicon”. Bacterial chromosome constitutes one replicon, whereas eukaryotic chromosomes contain numerous replicons. Replication proceeds either in one direction (unidirectional) or two directions (bidirectional) (Fig. 3-2). DNA replication is an enzymatic process involving DNA polymerases I and III, which require a primer (an oligonucleotide that is H-bonded to the template strand) with a 3’-OH group onto which a dNTP can attach. Chain elongation occurs only in the 5’→3’direction. (1) leading strand: synthesized continuously from 5’ to 3’ 2 (2) lagging strand: synthesized discontinuously (Fig. 3-3). Okazaki fragments can act as a primer. The nicks between fragments are sealed by DNA ligase. 9. DNA polymerase I has both 3’-5’and 5’-3’exonuclease activity (remove or excise nucleotides just added). This “proofreading” function ensures fidelity of replication. 10. Replication may be divided into three stages: (1) Initiation (2) Elongation: the complex of proteins associated with this stage is called a “replisome ”. (3) Termination DNA transcription 1. Transcription: the RNA molecule is synthesized on a DNA template by an enzymatic process that involves several distinct events. (1) Transcription starts at a specific site to which the appropriate enzyme binds. (2) Then polymerization is initiated, followed by chain elongation. (3) Finally, chain termination occurs and RNA molecule is released. (4) Promoters: binding sites on the DNA molecule. These are base sequences of about 40 bp in length. (5) Only one DNA strand is the template strand (antisense strand). The other strand is the sense or coding strand (Fig. 3-4). (6) RNA polymerase must recognize and bind to the promoter before transcription. 2. The product of transcription: messenger RNA (1) The term “cistron” is used to refer to a segment of DNA corresponding to a polypeptide, including the start and stop sequences. (2) When an mRNA codes for one polypeptide, it is called a “monocistronic mRNA”. (3) Frequently, prokaryotic mRNA is polycistronic, encoding several different polypeptide chains. (4) Polycistronic mRNA may be interspersed by sequences called “spacers”. (5) The DNA in the chromosomes of eukaryotes is tightly bound to nucleoproteins (histones), forming a complex structure called “chromatin”. (6) Transcription in eukaryotes occurs in the nucleus, and then the mRNA is transported out of the nucleus into the cytoplasm for translation. 3. Regulation of transcription (1) Two types of regulatory sequences located upstream from the point of initiation are involved in the stimulation and initiation of gene transcription in eukaryotes. (2) Specific proteins called “transcriptio nal factors” that facilitate the binding of RNA polymerase II recognize these sequences (promoters and enhancers). (3) Most promoters have a sequence comprised of repeats of thymine (T) and adenine (A) nucleotides (called the TATA box) that is usually located about -25 bp upstream of 3 the start point. (4) There are other promoter modules such as the CCAAT and GC boxes. (5) Promoters and their associated transcriptional factors control the degree of transcription initiation and consequently the amount of transcription of the gene. (6) Enhancers are the sequences that increase the transcriptional activity of genes. They interact with promoters to increase the rate of transcription initiation. (7) Enhancers vary in their location, and may be found upstream, downstream, or even within the gene. Whereas a promoter must be in a relatively fixed location with regard to the start point of transcription. 4. Post-transcriptional processing (1) The initial product of transcription in eukaryotes is a pre- mRNA, which requires considerable processing to produce a native mRNA. (2) Most eukaryotic genes are interspersed by non-coding sequences called “introns ” (i.e., the coding sequences are called “exons ”). (3) Introns are removed by the process called “splicing” in more than one way, consequently yielding different collections of exons in the mature mRNA. This mechanism is called “alternative splicing” and yields different but related proteins called “isoforms ” upon translation (Fig. 3-5). 1. 2. 3. 4. 5. 6. 7. 8. 9. Translation of mRNA (Peptide synthesis) Translation is a process by which mRNA is converted into an amino acid sequence and then polymerized into a polypeptide chain and eventually, protein. A triplet of bases called a “codon” encodes an amino acid. The collection of triplet codons that specify specific amino acid is called the “genetic code” (Fig. 3-6). The reading of the genetic code begins at a fixed point in the gene and proceeds sequentially without any interruption (reading frame). When there is an interruption by a mutation (insertion or deletion), the consequence is a shift in the reading frame (causing a frameshift mutation) (Fig. 3-7). Stop codons: UAA, UAG, and UGA, signal codes for termination of translation. Start codon: AUG, signal for the initiation of a polypeptide chain The genetic code is universal. Peptide synthesis in prokaryotes is as follows: (1) Initiation a. The initiation of translation involves a 30S (small ribosome subunit), an mRNA molecule, a specific charged initiator tRNA, guanosine 5’-triphosphate (GTP), Mg2+, and ar least three proteins called “initiator factors”. b. The linking of a tRNA to its respective amino acid is called “charging”. c. The tRNA so linked is said to be “charged” or “acylated” (because the enzyme 4 involved in charging is aminoacyl tRNA synthetase). d. Wobble hypothesis: Proposed by Francis Crick to describe that only the first two bases of a codon are critical to coding for a specific amino acid. e. The initiation codon (AUG) binds to an initiator tRNA that is charged into a formylated amino acid called “formylmethionine” to form fMet-tRNA. f. The binding involves a unique sequence of AGGAGGU (Shine-Dalgarno sequence) that occurs near the initiation codon. g. Shine-Dalgarno sequence forms base pairs with a complementary region of the 16S rRNA of the small ribosome. h. The fMet-tRNA binding is facilitated by another initiation factor. i. The resulting 30S preinitiation complex is jointed by a 50S subunit to produce a “70S initiation complex” (Fig. 3-8). (2) Elongation a. The 50S subunit contains two sites: P (peptidyl) site and A (aminoacyl) site. b. Peptidyl transferase catalyzes the formation of a peptide bond between two amino acids (Fig. 3-9). c. Translocation: once the peptide bond is formed, the P-site tRNA leaves the ribosome while the A site tRNA moves to the P site. d. The release and shifting of the entire mRNA-tRNA-aa2 -aa1 complex is facilitated by a number of protein called “elongation factors”. e. In E. coli, the elongation rate is about 15 amino acids per sec at 37o C, and the error rate is about 10-4 . (3) Termination a. Elongation is brought to an end when a stop codon is encountered. b. The stop codons prevent the A site from being occupied by tRNA and cause GTP-dependent proteins “release factors” to act to cleave the polypeptide chain. c. mRNA is also degraded. 10. Various parts of the mRNA may be translated simultaneously, each with its ribosome complex. The resulting structure is called a “polyribosome” or “polysome”. Cellular metabolism 1. Metabolism (1) Anabolism: the synthesizing reactions occur in the cell. (2) Catabolism: the destructive reactions occur in the cell. 2. Photosynthesis (1) This assimilation process occurs in the chloroplasts, which are found mainly in leaves. (2) These are plastids that contain the green pigment “chlorophyII” (an enzyme). a. ChlorophyII a occurs in all photosynthesizing eukaryotes. 5 b. ChlorophyII b occurs in vascular plants and bryophytes. (3) General reaction for photosynthesis: 6CO2 + 12H2O → C6 H12 O6 + 6O2 + 6H2O (4) C3 pathway (Calvin cycle) (Fig. 3-11). (5) C4 pathway. Cellular respiration 1. The process by which active cells obtain energy from food. 2. Occurs in mitochondria. 3. May be described as the reverse of photosynthesis. 4. Usually requires oxygen to occur and is thus called “aerobic respiration”. 5. Under certain conditions, it may occur in an oxygen-deficient environment and is called “anaerobic respiration”. 6. General reaction for aerobic respiration: C6 H12 O6 + 6O2 → 6CO2 + 6H2 O + energy 7. There are three stages in aerobic respiration: (1) Glycolysis (Fig. 3-13). a. Glucose is converted to pyruvic acid. b. 2 ATP is obtained. (2) Kreb’s (tricarboxylic acid) cycle (Fig. 3-14). a. also called “oxidative decarboxylation”. (3) Electron transport chain (Fig. 3-15). a. Electron transport involves the flow of electrons in an energetically downhill fashion, resulting in energy release for the formation of ATP by a process called “oxidative phosphorylation”. 8. At the end of one cycle of aerobic respiration, one molecule of glucose yields a net of 36 ATPs. 9. The end product of anaerobic respiration is ethyl alcohol (ethanol). This process is also called “fermentation” (Fig. 3-16). 10. Only two ATP molecules are obtained per molecule of glucose in anaerobic respiration. Key concepts Websites tutorial See p. 46. References Buratowski, S. 1995. Mechanisms of gene activation. Science, 270: 1773-1774.