* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download •MOLECULAR CELL BIOLOGY
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Genealogical DNA test wikipedia , lookup
United Kingdom National DNA Database wikipedia , lookup
Bisulfite sequencing wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Cancer epigenetics wikipedia , lookup
DNA damage theory of aging wikipedia , lookup
Human genome wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Gel electrophoresis of nucleic acids wikipedia , lookup
Nutriepigenomics wikipedia , lookup
RNA interference wikipedia , lookup
DNA polymerase wikipedia , lookup
Designer baby wikipedia , lookup
DNA vaccination wikipedia , lookup
Molecular cloning wikipedia , lookup
Short interspersed nuclear elements (SINEs) wikipedia , lookup
Cell-free fetal DNA wikipedia , lookup
Epigenomics wikipedia , lookup
Extrachromosomal DNA wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Point mutation wikipedia , lookup
Nucleic acid double helix wikipedia , lookup
DNA supercoil wikipedia , lookup
Microevolution wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
History of genetic engineering wikipedia , lookup
Polyadenylation wikipedia , lookup
Messenger RNA wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Nucleic acid tertiary structure wikipedia , lookup
RNA silencing wikipedia , lookup
Non-coding DNA wikipedia , lookup
Helitron (biology) wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
History of RNA biology wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Epitranscriptome wikipedia , lookup
Non-coding RNA wikipedia , lookup
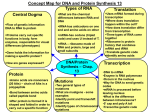
Nucleic acid (macro-molecules): Determining the correct order amino acids sequence → structure and function One ribosomal RNA transcription Lodish • Berk • Kaiser • Krieger • Scott • Bretscher •Ploegh • Matsudaira • MOLECULAR CELL BIOLOGY • SIXTH EDITION • CHAPTER 4 • Basic Molecular Genetic Mechanisms ©Copyright 2008 W. H.©Freeman andand Company 2008 W. H. Freeman Company Gene (DNA) contains all information → build the cells and tissues of organism Deoxyribonucleic acid (DNA) contains the information prescribing the amino acid sequence of proteins This information is arranged in units termed genes Ribonucleic acid (RNA) serves in the cellular machinery that chooses and links amino acids in the correct sequence The central dogma: DNA ⌫ RNA ⌫ Protein DNA and RNA are polymers of nucleotide subunits RNA + ribose + protein → ribosomal ribonucleoprotein complexes (rRNPs) Chemical structure of the principal bases (ch3) Monomers→polymers DNA: ATCG RNA: AUCG Four basic molecular genetic processes: Protein synthesis: 1 to 3 rRNA: ribosomal RNA; tRNA: transfer RNA rNTPS: ribonucleoside triphosphate monomers; dNTP:deoxyribonucleoside triphophate 專有名詞要不止要 放在心中,更要放 在腦中 Structure of nucleic acid A nucleic acid strand is a linear polymer with end to end directionality REMEMBER: DNA = deoxyribonucleotides; RNA = ribonucleotides (OH-groups at the 2’ position) Note the directionality of DNA (5’-3’ & 3’5’) or RNA (5’-3’) DNA = A, G, C, T ; RNA = A, G, C, U Nucleotide subunits are linked together by phosphodiester bonds Native DNA is a double helix of complementary antiparallel strands 1953, Watson and Francis: proposed that DNA has a double-helical structure Nature, 4356, 737-728 (1953) DNA consists of two associated polynucleotide strands that wind together to form a double helix. 5’→3’; 3’→5’ antiparallel Base pair: H-bond formation, A-T (2) and G-C (3) Complementary: two polynucleotide consequence of the size, shape and chemical composition, by base pair interaction (A-T and C-G) There are two major forces that contribute to stability of helix formation: Hydrogen bonding in base-pairing Hydrophobic interactions in base stacking (堆) Nucleic acid as hetero-polymers Nucleosides, nucleotides (Ribose sugar, (2’-deoxy ribose sugar, RNA precursor) DNA precursor) (2’-deoxy thymidine triphosphate, nucleotide) DNA and RNA strands So … DNA RNA Most DNA in cells is a right handed helix X-ray data of DNA: (B-form) 1.The stacked bases are regularly spaced 0.34-0.36nm 2.Helix makes a complete turn every 3.6nm, about 10.5 pairs per turn. B-DNA A-DNA B DNA most common d(CGCGAATTCGCG)•d(CGCGAATTCGCG) A DNA, in low humidity condition, B transform to A form; RNA-RNA, RNA-DNA d(AGCTTGCCTTGAG)•d(CTCAAGGCAAGCT) Z DNA, short DNA molecules composed of alternating purine-pyrimidine nucleotides (GC), right transform to left d(CGCGCGCGCGCG)•d(CGCGCGCGCGCG) Z-DNA DNA compositional biases B-DNA A-DNA R.H. helix R.H. helix Z-DNA Base compositions of genomes: G+C (and therefore also A+T) content varies between different genomes The GC-content is sometimes used to classify organism in taxonomy High G+C content bacteria: Actinobacteria e.g. in Streptomyces coelicolor it is 72% 鏈黴菌 Low G+C content: Plasmodium falciparum (~20%) 瘧原虫 Other examples: Saccharomyces cerevisiae (yeast) 38% Arabidopsis thaliana (plant) 36% Escherichia coli (bacteria) 50% L.H. helix TBP protein can binds to the minor groove of specific DNA (rich AT)→ untwisting and sharply bending the double helix → transcription ability ↑ Why is rich AT region ? Why RNA degradation more easy than DNA??? DNA RNA Base-catalyzed hydrolysis of RNA 2’-OH site as a nucleophile at normal pH → attacking phophodiester bond→ degradation In 2’ site, DNA is more stable than RNA. DNA can undergo reversible strand separation (denaturation) Tm: melting temperature G-C more → need more energy Denature of single stranded DNA → random coil (without organized structure) Renature vs hybribization SV40 viral DNA Many prokaryotic genomic DNA and viral DNA are circular molecules. Circular DNA molecules in eukaryotic mitochondria and chloroplasts Circular DNA without end, when replication: open DNA → unwinding DNA → torsional (扭 力) stress → winding (纏繞) → formed super-coil (超螺旋) Topoisomerase I (bacterial and eukaryotic cell has) → bind to DNA → breaks a phosphodiester bond in one strands DNA formed nick → loss supercoiled → ligates the two ends of the broken strand. Topoisomerase II, breaks two strands DNA Supercoils Supercoiling of DNA can only occur in closed-circular DNA or linear DNA where the ends are fixed. Underwinding produces negative supercoils, whereas overwinding produces positive supercoils. Supercoiling induced by separating the strands of duplex DNA (eg., during DNA replication) DNA (double strain) → open → single strain → replication or transcription→ spuercoiling → need topoisomerase Transcription of protein-coding genes and formation of functional mRNA Relaxed and supercoiled plasmid DNAs DNA → RNA → Protein → function ATCG AUCG mRNA tRNA rRNA Encode: AA protein –coding gene gene → mRNA → protein DNA replication Direction 5’ to 3’ ~800 nd/sec RNA polymeration: Direction 5’ to 3’ ~40 nd/sec Translation Direction 5’ to 3’ ~15 aa/sec Different types of RNA exhibit various conformations related their functions AUCG: CG has 3 H-bond Most RNA are single strand Various RNA → carry out specific functions Eukaryotic cell, RNA self-splicing > Secondary structure H-bond dependent 5-10 nucleotides >10 nucleotides Three Different Classes of RNA 1) rRNA (ribosomal) • large (long) RNA molecules • structural and functional components of ribosomes • highly abundant 2) mRNA (messenger) • typically small (short) • encode proteins • multiple types, not abundant 3) tRNA (transfer) and small ribosomal RNAs • very small • Important in translation Not all genes encode proteins DNA DNA Deoxyribonucleic acid ATCG More rigid More stable RNA Ribonucleic acid AUCG More flexible More unstable mRNA, rRNA, tRNA transcription RNA A template DNA strand is transcribed into a complementary RNA chain by RNA. Ribonucleoside triphosphate (rNTP) are polymerized to form a complementary RNA by RNA polymerase. Polymerization involves a nucleophilic attack by the 3’ oxygen in the growing RNA chain on the a phosphate of the next nucleotide → formed phosphodiester bond and release pyrophosphate Direction: 5’→ 3’; opposite in polarity to their template DNA strands DNA A→U T→A C→ G G→C transcribed to RNA Release PPi The micro RNA (miRNA): 1. Regulate specific mRNA 2. Produced by RNA polymerase RNA polymerase begins transcription is +1 Downstream: +, Upstream: - pri-miRNA :由基因組中轉錄出來 Drosha :一種RNaseIII pre-miRNA:由Drosha切割pri-miRNA 而來 Exportin-5 :可將pre-miRNA由細胞核運到 細胞質。 miRNA duplex : pre-miRNA被切割後的產物 (20~22個鹼基 ) mature miRNA:有活性的單鏈miRNA Bacterial (Prokaryotic) Transcription Three stages in transcription Promoters - DNA sequences that guide RNA polymerase to the beginning of a gene (transcription initiation site). Terminators - DNA sequences that specify then termination of RNA synthesis and release of RNAP from the DNA. Need transcription factor help Many transcription factor binding for help RNA polymerase binding RNA Polymerase (RNAP) - Enzyme for synthesis of RNA. Reaction (ordered series of steps) 1) Initiation. 2) Elongation. 3) Termination. About 14 base pairs Recognition rNTP vs.dNTP Three stages in transcription Termination of transcription Two mechanisms 1) Rho - the termination factor protein 初期 About 8 base pair For continuous RNA synthesis and without dissociation – rho is an ATP-dependent helicase – it moves along the RNA transcript, finds the "bubble", unwinds it and releases the RNA chain. 2) Rho-Independent - termination sites in DNA inverted repeat, rich in G:C, which forms a stem-loop in RNA transcript Rho-Dependent Transcription Termination (depends on a protein AND a DNA sequence) G/C -rich site Termination of transcription Two mechanisms 2) Rho-Independent - termination sites in DNA RNAP slows down Rho helicase catches up Elongating complex is disrupted – inverted repeat, rich in G:C, which forms a stem-loop in RNA transcript Rho-independent transcription termination Rho-Independent Transcription Termination (depends on DNA sequence - NOT a protein factor) • RNAP pauses when it reaches a termination site. • The pause may give the hairpin structure time to fold • The fold disrupts important interactions between the RNAP and its RNA product • The U-rich RNA can dissociate from the template Stem-loop structure DNA A T G C Transcriptional mechanism-1 Bacterial RNA polymerase RNA T • The complex is now disrupted U and elongation is terminated C G RNA Polymerase Structure of RNA polymerase • RNA polymerase are similar in eukaryotic and prokaryotic cell Five subunit: 2 large subunit: β, β’; 2 smaller subunits α and ω (only Stabilizes and assembly of its subunits) Only a single RNA polymerase (prokaryotic) In E.coli, RNA polymerase is 465 kD complex, with 2 α, 1 β, 1 β', 1 σ β' binds DNA 2+ β binds rNTPs and interacts withMg σ β and β ' together make up the active site α subunits appear to be essential for assembly and for activation of Current model of bacterial RNA enzyme by regulatory proteins polymerase bond to a promoter Different Types of RNA Polymerase In Bacteria (simple system) - all three classes are transcribed by the same RNA polymerase (RNAP for short) In Eukaryotes (complex system) - each class is transcribed by a different RNA Polymerase •RNAP I - rRNAs •RNAP II - mRNAs •RNAP III - tRNAs & small ribosomal RNAs •Remember: only RNAP did not transcript !!!! Need many transcription factor (protein) FlashFlash-2 1 2 3 4 5 RNA Polymerase is a spectacular (壯觀) enzyme, functioning in: Recognition of the promoter region Melting of DNA (Helicase + Topisomerase); unwinding DNA RNA Priming (Primase) RNA Polymerization; add rNTP Recognition of terminator sequence RNA-DNA hybrid Length? 3 to 9 bases, it is short and transit In Bacterial which can hold~16 bp In yeast which can hold ~25 bp Thus, RNAP is a multisubunit enzyme One model for transcriptional activation Gene Regulation Protein complex → DNA → open/tight DNA → transcription Transcription is regulated by proteins binding to or near promoters – Three types of proteins involved: • Specificity factors • Repressors • Activators – Repressors: bind to specific sites on DNA • Called operators • Either near or overlapping the promoter • Block movement of RNA-polymerase -Activators: bind to specific sites on DNA, help RNAP moving Operon: arrangement of genes in a functional group Organization of genes differs in prokaryotic and eukaryotic DNA Genomes In prokaryotic: 1. logic: genes devoted (致力於) to a single metabolic goal; protein synthesis from a contiguous array in DNA. It means that one gene → one protien→. 可以多段有功能基因連在一起 one operon → one goal (function) 2. Arrangement of genes in a functional group is cell an operon, because it operate as a unit from a single promoter. One promoter → one gene ( or genes) → one protein (or proteins) 3. The genes are closely packed with very few non-coding gaps DNA → direct to co-linear mRNA → → translated protein Eukaryotic precursor mRNA are processed to form functional mRNAs In eukaryotic 1. RNA → discontinuous in corresponding DNA sequence 2. DNA contain exons (coding sequence) and introns (nonprotein-coding segments) 3. DNA → RNA, remove introns and carefully stitched back together to produced many mRNAs 4. Functional (mature) mRNA from precursor mRNA processed (splicing) 5. DNA → pre-mRNA → splicing → mature mRNA→ protein → add modification → mature protein Each gene is transcripbed from its own promoter Tryptophan (trp) Tryptophan metabolite enzyme In prokaryotic, protein synthesis can occur in 5’ or 3’ end of mRNA; transcription and translation can occur at the same time. In eukaryotic, in nucleus DNA → transcription → precursor mRNA → procession → functional mRNA → transport to cytoplasm → translated to protein; Transcription and translation are in different time and place. Pre-mRNA are modified at the tow ends, and keep in mRNA. It can protect the degradation of RNA form nucleus to cytoplasm. Don’t need DNA template. Modification of 5’ end: by RNA polymerase II → add 5’cap; methylation Modification of 3’ end: by poly A polymerase, add 100-250 A and produced poly A tail. mRNA processing – RNA splicing, 5’ and 3’ retain noncoding regions (untranslated regions; UTRs). In mammalian mRNA, 5’ UTR about >100 nucleotides, 3’ UTR about several kilobases The ribose of the second nucleotide also is methylated Alternative RNA splicing increase the number of proteins expressed from a single eukaryotic gene RNA Processing: • Prokaryotes: transcription and translation can be One gene → RNA splicing → different RNA→ different protein Isoform: by alternative splicing production of different forms of a protein. Untranslated region One gene can lead to more than one protein (e.q. antibodies) concurrent. • Eukaryotes: Nucleus (RNA synthesis) and cytoplasm (Protein synthesis) are separated. • Primary transcript undergoes several modifications. • 5’ cap is added to 5’ nucleotide; m7Gppp (Stability) Exons: part of the gene that is expressed. Introns: part of gene that is spliced out from pre-mRNA. • String of adenylic acids are added to the 3’ end (Poly A tail) Sometimes some exons are also spliced out. • Splicing: internal cleavage to excise introns followed by ligation of coding exons Formed three protein-coding exon 5’ and 3’ ends of eukaryotic mRNA Functions of 5’ cap and 3’ polyA Both cap and polyA contribute to stability of mRNA: – Most mRNAs without a cap or polyA are degraded rapidly. – Shortening of the polyA tail and decapping are part of one pathway for RNA degradation in yeast. Need 5’ cap for efficient translation: Add a GMP. Methylate it and 1st few nucleotides Cut the pre-mRNA and add A’s – Eukaryotic translation initiation factor 4 (eIF4) recognizes and binds to the cap as part of initiation. – Assists mRNA export to the cytoplasm 轉到ch6 p216 p249 Gene: DNA regions encoding proteins or functional RNA Intron: non-functional DNA, non-coding regions of DNA Extron: functional DNA, coding region of DNA Transposable (mobile) DNA: non-coding region, repeat, evolutionary DNA must be contend: human cell has 2 meters DNA!!!!!SO must be highly compacted In eukaryotes, DNA + protein → chromatin → chromosome histone The structure of genes and chromosomes Eukaryotic gene structure A gene: as the entire nucleic acid sequence that is necessary for the synthesis of a functional gene product Coding region: coding amino acids sequence, or functional RNA Enhancer: transcript regions, not coding region; it regulated transcriptive activity Most eukaryotic genes contain introns and produce mRNA encoding single proteins Simple and complex transcriptions units are found in eukaryotic cells Cistron: a genetic unit encoding a single polypeptide Polycistron: a genetic unit (not a only a gene) encoding multiple polypeptides; also called operon, like prokaryotic cell for live Most eukaryotic cell has mono-cistron. Prokaryotes have compact genomes and their transcripts often contain multiple protein coding regions (called open reading frames or ORFs) These mRNAs are called polycistronic mRNAs (a cistron is a concept that is similar to a gene, and for many genes the cistron=gene) Homologous recombination: meiosis Exon 3 is lost L: non-coding repeat, also called Transposable (mobile) DNA its easy to homologous recombination Homologous recombination and generate genetic diversity Generate genetic diversity among the individuals of a species by causing the exchange of large regions of chromosomes between the maternal and paternal pair of homologous chromosomes during the cellular division the generates germ cells Simple and complex eukaryotic transcription Mutation control region: no mRNA expression → no protein → no function Mutation Exon : mRNA expression (some wrong) → abnormal protein → activity change For gene that are transcribed from different promoters (regulator factor) in different cell type Protein-coding genes may be solitary or belong to a gene family Solitary gene: in multicellular organism, 20-50% protein coding gene are reprsented Duplicated gene: gene family → protein family homologous duplicated gene encode protein with similar New Roles of RNA RNAi - RNA interference siRNA- active molecules in RNA interference; degrades mRNA (act where they originate) miRNAs - tiny 21–24-nucleotide RNAs; probably acting as translational regulators of protein-coding mRNAs stRNA - Small temporal RNA; (ex. lin-4 and let-7 in Caenorhabditis elegans snRNA - Small nuclear RNA; includes spliceosomal RNAs (processing) snoRNA - Small nucleolar RNA; most known snoRNAs are involved in rRNA modification Alternative RNA splicing increases the number or proteins expressed from a single eukaryotic gene Production of heavy chain genes in mouse by recombination of V, D, J, and C gene segments during development Higher eukaryote have multidomain tertiary structure only from a small number of exons. Single gene →Multiple introns→alternative splicing → protein isoforms Alternative splicing: The presence of multiple introns in many eukaryotic genes permits expression of multiple, related proteins form a single gene. > 20 isoforms fibronectin from different alternatively spliced mRNA Cell type specific splicing of fibronectin pre-mRNA • Alternative splicing – Different mRNAs can be produced by same transcript Peter J. Russell, iGenetics: Copyright © Pearson Education, Inc., publishing as Benjamin Cummings. Differences Between Transcription In Prokaryotes and Eukaryotes Transcription And Translation In Prokaryote-----------the same time Eukaryotic Transcription and translation--------------different time Processing Eukaryotic mRNA