* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download This is going to be a long journey, but it is crucial
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Gene expression profiling wikipedia , lookup
Epigenomics wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Cell-free fetal DNA wikipedia , lookup
Short interspersed nuclear elements (SINEs) wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
RNA interference wikipedia , lookup
Designer baby wikipedia , lookup
History of genetic engineering wikipedia , lookup
Nucleic acid tertiary structure wikipedia , lookup
Transfer RNA wikipedia , lookup
Transcription factor wikipedia , lookup
RNA silencing wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Expanded genetic code wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Non-coding DNA wikipedia , lookup
Frameshift mutation wikipedia , lookup
Microevolution wikipedia , lookup
Polyadenylation wikipedia , lookup
Helitron (biology) wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
History of RNA biology wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Genetic code wikipedia , lookup
Non-coding RNA wikipedia , lookup
Point mutation wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Messenger RNA wikipedia , lookup
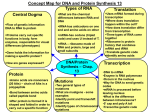
GUIDED READING - Ch. 17 - FROM GENE TO PROTEIN • NAME: ________________________ Please print out these pages and HANDWRITE the answers directly on the printouts. Typed work or answers on separate sheets of paper will not be accepted. • • • • Importantly, guided readings are NOT GROUP PROJECTS!!! You, and you alone, are to answer the questions as you read. You are not to share them with another students or work together on filling it out. Please report any dishonest behavior to your instructor to be dealt with accordingly. Get in the habit of writing legibly, neatly, and in a NORMAL, MEDIUM-SIZED FONT. AP essay readers and I will skip grading anything that cannot be easily and quickly read so start perfect your handwriting. Please SCAN documents properly and upload them to Archie. Avoid taking photographs of or uploading dark, washed out, side ways, or upside down homework. Please use the scanner in the school’s media lab if one is not at your disposal and keep completed guides organized in your binder to use as study and review tools. READ FOR UNDERSTANDING and not merely to complete an assignment. Though all the answers are in your textbook, you should try to put answers in your own words, maintaining accuracy and the proper use of terminology, rather than blindly copying the textbook whenever possible. “This is going to be a long journey, but it is crucial to your understanding of biology. Work on this chapter a single concept at a time,” [2] review often, and take your time trying to understand the processes discussed. 1. What is ‘gene expression’? Genes specify proteins via transcription and translation [2]. 2. Who was Archibald Garrod and what did he refer to as ‘inborn errors of metabolism’? 3. Describe one example Garrod used to illustrate his hypothesis. [2] 4. State the hypothesis formulated by George Beadle while studying eye color mutations in Drosophila. [2] 5. a. Beadle and Tatum set out to study if individual genes specify the enzymes that function in biochemical pathways. Which organism did Beadle and Tatum use in their research? b. How did this organism’s nutritional requirement facilitate this research? [2] c. How were Neurospota spores treated to increase the mutation rate? [2] d. What is the difference between a minimal medium and a complete growth medium? e. How did they identify only mutants incapable of synthesizing arginine? f. Describe the Beadle and Tatum experiment with the bread mold, Neurospora crassa, in DETAIL. As you do so fill in the diagram below to accompany your explanation. The logic behind the experiment, the interpretation of its results, and the conclusion drawn are critical! [3] Classes of Neurspora crassa 6. What were two significant findings that resulted from the research of Beadle and Tatum? [2] 7. What was Beadle and Tatum’s final hypothesis? 8. What later revisions to the one gene-one enzyme hypothesis were necessary as more information was gained? A critical concept! Basic principles of transcription and translation [2]. 9. What are three ways in which RNA differs from DNA? [2] a. b. c. 10. What are the monomers of DNA and RNA? [2] 11. What are the monomers of proteins? [2] 12. Define the following processes that are part of the making of proteins. a. Transcription b. Translation 13. What is Messenger RNA or mRNA? 14. What is a ribosome? 15. How is pre-mRNA different from mRNA? 16. In eukaryotes, what is the pre-mRNA called? 17. Complete the following table to summarize the processes of transcription and translation. [2] 18. What is the Central Dogma of molecular genetics, as proclaimed by Francis Crick? [2] 19. a. How many nucleotide bases are there? [2] b. How many amino acids are there? [2] 20. Label the flow of genetic information in a prokaryotic and eukaryotic cell. 21. Why is the genetic code a triplet code of non-overlapping nucleotides as opposed to a single or double base one? 22. So, how many nucleotides are required to code for these 20 amino acids? 23. How many unique triplet codes exist? 24. What is the ‘template strand’? 25. In what direction is the RNA molecule synthesized during DNA transcription? 26. a. Here is a short DNA template. Below it, assemble the complimentary mRNA strand. (*Note that you MUST always identify the 3’ and 5’ ends of any nucleic acid polymer written! It is convention to write the 5’ end first so pay attention to orientation first before answering any question.) 3’ A C G A C C A G T A A A 5’ b. How many codons are there above? c. Label one codon. d. What would be the nontemplate strand of the gene (the DNA coding strand)? Label the 3’ and 5’ ends of course. e. How does the nontemplate DNA strand compare to the mRNA polymer? [1] f. Imagine that the nontemplate sequence (aka the coding sequence) would be transcribed instead of the template strand. Draw the mRNA sequence that would result. Label the 3” and 5” ends of course. [1] g. The ribosome reads the mRNA starting from the 5’ end! Now translate this mRNA sequence made by transcribing the nontemplate DNA strand. [1] h. How well do you think the protein synthesized from the nontemplate strand would function? [1] 27. How did Nirenberg “figure out” which amino acids was specified by which code? 28. What was the first codon-amino acid pair to be identified? [2] 29. What polypeptide product would you expect form a poly-G mRNA that is 30 nucleotides long? [1] 30. a. What is the mRNA sequence of the start codon? b. What amino acid does it code for? c. Of the 64 codons, how many of the codons result in an amino acid being added to a growing polypeptide chain? Explain. d. What are the sequences for the three stop codons? e. What actually happens when the ribosome hits a stop codon in the mRNA? 31. What is meant by the statement “there is redundancy in the genetic code, but no ambiguity”? 32. What is the reading frame? [3] 33. DNA is DNA is DNA. Meaning that the code is nearly universal. Because of this jellyfish genes can be inserted into pigs, or firefly genes can make a tobacco plant glow! [2] What conclusions can be drawn from the near universality of the genetic code of all living organisms? [3] Transcription is the DNA-directed synthesis of RNA: A closer look [2]. 34. Which is the enzyme that uses the DNA template strand to transcribe a new mRNA strand? [2] 35. Recall that DNA pol III adds new nucleotides to the template DNA strand to assemble each new strand of DNA. Both enzymes can assemble a new polynucleotide in the 5’ ! 3’ direction. Which enzyme, DNA polymerase III or RNA polymerase does not require a primer to begin synthesis? [2] 36. What is a transcription unit? 37. What is the promoter? 38. What are meant by the terms ‘downstream’ and ‘upstream’? 39. a. What is the transcription start point and where is it located? b. Is the transcription start point upstream or downstream of the transcription unit? [1] 40. Label the diagram illustrating the three stages of transcription. To the right of the figure, name the three stages of transcript and briefly describe each stage. [2] 41. Let’s now take a closer look at initiation. Read the paragraph titled “RNA Polymerase Binding and Initiation of Transcription” carefully. List three important facts about the promoter here. [2] 1. 2. 3. 42. Use the diagram below to demonstrate initiation of transcription at a eukaryotic promoter. Also label the 5’ and 3’ ends of the DNA and mRNA. To the right of the figure, explain the three stages of initiation that are shown. TATAAAA ATATTTT 43. What are transcription factors? 44. What is the difference then between the binding of RNA polymerase to the promoter in prokaryotes like bacteria and eukaryotes like humans? 45. a. What is the TATA box found in eukaryotes and what is its function in initiating transcription? [2] b. How do you think it got this name? [2] 46. Suppose X-rays caused a sequence change in the TATA box of a particular gene’s promoter. How would that affect alter initiation and finally the transcription of the gene? [1] 47. What comprises a transcription initiation complex? [2] 48. Contrast the termination of transcription in prokaryotes and eukaryotes. 49. Now its time to put al of the elements of transcription together. Write an essay to describe the process by which mRNA is formed IN EUKARYOTES. Use the following terms correctly in your essay and underline each: TATA box, gene, terminator, promoter, elongation, 5’ to 3’, termination, initiation, RNA polymerase, RNA nucleotides, template, start point, termination signal, and transcription factors. This is a typical essay on the AP Bio exam. [2] _____________________________________________________________________________________________________ _____________________________________________________________________________________________________ _____________________________________________________________________________________________________ _____________________________________________________________________________________________________ _____________________________________________________________________________________________________ _____________________________________________________________________________________________________ _____________________________________________________________________________________________________ _____________________________________________________________________________________________________ _____________________________________________________________________________________________________ _____________________________________________________________________________________________________ _____________________________________________________________________________________________________ _____________________________________________________________________________________________________ _____________________________________________________________________________________________________ _____________________________________________________________________________________________________ _____________________________________________________________________________________________________ _____________________________________________________________________________________________________ _____________________________________________________________________________________________________ _____________________________________________________________________________________________________ _____________________________________________________________________________________________________ _____________________________________________________________________________________________________ _____________________________________________________________________________________________________ _____________________________________________________________________________________________________ _____________________________________________________________________________________________________ _____________________________________________________________________________________________________ _____________________________________________________________________________________________________ _____________________________________________________________________________________________________ _____________________________________________________________________________________________________ _____________________________________________________________________________________________________ _____________________________________________________________________________________________________ _____________________________________________________________________________________________________ _____________________________________________________________________________________________________ _____________________________________________________________________________________________________ _____________________________________________________________________________________________________ _____________________________________________________________________________________________________ _____________________________________________________________________________________________________ _____________________________________________________________________________________________________ Eukaryotic cells modify RNA after transcription [2]. 50. Recall that eukaryotes modify pre-mRNA before the final mRNA is ready to be sent to the cytoplasm for translation. What three alterations occur during this RNA processing? Be specific about what happens during each modification and where in the pre-mRNA transcript the modification occurs. 1. 2. 3. 51. What are the three important roles of the 5’ cap and poly-A tail added to the ends of the mRNA transcript? 52. What are the 5’ and 3’ UTR’s and what is their function? 53. Label the diagram below showing the mRNA that results after RNA Processing. G-P-P-P AAUAAA 54. What is RNA Splicing? 55. Distinguish between introns and exons. Remember Exons are Expressed! AAAAAAAA… 56. Label the diagram showing the results of RNA Splicing. 57. a. What are snRNPs? [2] b. What two types of molecules make up a snurp? (Snurps not smurfs! ) [2] 58. You will be introduced to a number of small RNAs in this course. What type is the RNA in a snRNP? [2] 59. Snurps band together in little snurp groups to form spliceosomes. How do spliceosomes work? [2] 60. On the figure below, label the following: pre-mRNA, snRNPs, snRNA, protein, spliceosomes, intron, exon, and other proteins. 61. Study the figure above and text carefully to explain how the splice sites are actually recognized. [2] 62. a. What is a ribozyme? b. What commonly held idea was rendered obsolete by the discovery of ribozymes? [2] c. What are three properties of RNA that allow it to function as an enzyme? [2] 1. 2. 3. 63. What is alternative RNA splicing of identical mRNA transcripts and why is it important? 64. What are domains and how to they relate to exons? 65. What is exon shuffling and what may be the benefit of it? Translation is the RNA-directed synthesis of a polypeptide: A closer look [2]. 66. Describe the structure and function of transfer RNA & label the molecule. 67. Compare and contrast the codon and anticodon? 68. a. Why is the enzyme aminoacyl-tRNA synthetase important to translation and protein synthesis? b. What are the three substrates and the main product of this enzyme? c. How many types of aminoacyl-rRNA synthetase enzyme does the cell have and why? d. How many types of tRNA does the cell have? e. How does wobble explain that there are fewer tRNAs than codons? f. To review, use the figure below to explain the process of a specific amino acid being joined to a tRNA in your own words. Use correct terminology and then also label the figure with aminoacyl-tRNA synthetase, ATP, amino acid, and tRNA. [2] 69. a. Describe the composition and structure of ribosomes. b. Where/how are the two components of the ribosome synthesized? c. Where are these components of the ribosomes assembled into ribosomal subunits? 70. a. How does a prokaryotic ribosome differ from a eukaryotic ribosome? [2] b. What is the medial significance of this difference? [2] 71. To review, three types of RNA are thus needed for protein synthesis. Complete the chart below. [2] 72. On this figure, label the large subunit, small subunit, A site, P site, E site, and mRNA binding site. [2] 73. Describe the following parts of the ribosome. a. P site b. A site d. E site e. Exit tunnel f. mRNA binding site 74. Much like transcription, we can divide translation into three stages. List them below. [2] 1. 2. 3. 75. Detail the steps of initiation of translation and label the diagram below. [3] 76. What is always the first amino acid in the new polypeptide? [2] 77. Use the diagram below to detail elongation cycle of translation. [3] 78. a. What are polyribosomes? b. Why are they beneficial to a cell? [1] c. Do they exist in eukaryotes or prokaryotes? Why? 79. What is a release factor and how does it aid translation termination? 80. Use the diagram below to detail the termination of translation. [3] 81. What begins to happen to the growing polypeptide as it exits the ribosome as far as shape is concerned? 82. List a few examples of a post-translational modification of a protein? 1. 2. 3. 4. 83. What is a ‘signal peptide’? 84. What is a ‘signal recognition particle’? 85. Proteins intended to be included in membranes of the endromembrane system or secreted from the cell head to the Rough Endoplasmic Reticulum. Once inside the ER, proteins may be processed further. This processing may even continue in the Golgi. First, however, proteins must enter the ER. Label the following illustration showing the signal mechanism for targeting proteins to the ER and explain the 6 steps in the boxes below. 1. 2. 3. 4. 5. 6. Point mutations can affect protein structure and function [2]. Mutations are the ultimate and original source of new alleles and genes. We studied gross (large-scale) mutations in chromosome structure already. Now you must understand more microscopic changes too. It is imperative that you thoroughly understand Figure 17.23 on page 345. 86. What is a mutation in terms of molecular genetics? 87. What are point mutations? 88. a. What type of point mutation is a base-pair substitution? b. What type of base-pair substitution is a silent mutation? c. What type of base-pair substitution is a missense mutation? d. What type of base-pair substitution is a nonsense mutation? 89. What type of point mutation is an insertion? 90. What type of point mutation is a deletion? 91. a. What are frameshift mutations? b. Identify two mechanisms by which frameshifts may occur. 92. Though a gene is hundreds to thousands of nucleotides long, suppose a gene whose template strand contains the sequence 3’-TACTTGTCCGATATC-5’ is mutated to 3’-TACTTGTCCAATATC-5’. This now comprises a new allele of the gene in a population if it is passed down in the gamete to the next generation. [1] a. For both normal and mutant genes, draw the double-stranded DNA, the resulting mRNA, and the amino acid sequence each encodes side by side below. b. What is the effect of the mutation on the amino acid sequence. c. What type of a mutation was this? 93. a. What are chemical and physical ‘mutagens’? b. What is an example of a physical mutagen? c. Describe the action of different types of chemical mutagens. [2] Although gene expression differs among the domains of life, the concept of a gene is universal [2]. 94. Describe two important ways in which bacterial and eukaryotic gene expression differ. [2] 95. Multiple choice: This illustration shows coupled transcription and translation in bacteria. [1] a. Which one of the mRNA molecules started transcription first (a. – d.)? b. Which ribosome started translating first (e. – j.)? DNA Gene Polyribosome a. b. c. d. e. f. g. h. i. 5’ end of mRNA 96. a. j. How has a gene been “redefined” from the simply stated one gene codes for one polypeptide to the broader molecular definition in use today? b. Why prompted this redefinition? 97. Use the diagram below to help you study the “whole” picture. See if you can label without looking at your text[2] 98. Please answer the Self-Quiz at the end of your chapter. Do your best to try it from memory first in order to test how well you grasped the material. 1. ______ 2. ______ 3. ______ 4. ______ 5. ______ 6. ______ 7. ______ References 1. Campbell et al. (2008). AP* Edition Biology. 8th Ed. San Francisco: Pearson Benjamin Cummings. 2. Adapted from Fred and Theresa Holtzclaw 3. Adapted from L. Miriello