* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download first of Chapter 11: Gene Regulation
Genomic imprinting wikipedia , lookup
Epigenetics in learning and memory wikipedia , lookup
Minimal genome wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Genome (book) wikipedia , lookup
Ridge (biology) wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Nucleic acid tertiary structure wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Oncogenomics wikipedia , lookup
RNA interference wikipedia , lookup
Gene expression programming wikipedia , lookup
Designer baby wikipedia , lookup
Transcription factor wikipedia , lookup
Genome evolution wikipedia , lookup
Non-coding DNA wikipedia , lookup
Polyadenylation wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
History of RNA biology wikipedia , lookup
RNA silencing wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Frameshift mutation wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Short interspersed nuclear elements (SINEs) wikipedia , lookup
Microevolution wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Gene expression profiling wikipedia , lookup
Messenger RNA wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Point mutation wikipedia , lookup
Non-coding RNA wikipedia , lookup
Primary transcript wikipedia , lookup
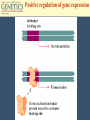
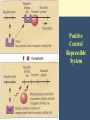
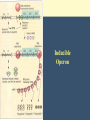
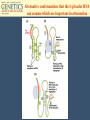
Chapter 11 Molecular Mechanisms of Gene regulation Jones and Bartlett Publishers © 2005 Categories of Protein-Coding Genes in Arabidopsis Regulation of Gene Expression Transcriptional RNA processing Translational mRNA stability Posttranslational control DNA rearrangements Prokaryotic transcriptional regulation • How ‘off’ is off? • Coordinate regulation Negative / Inducible / Repressible Positive regulation of gene expression Negative Control Inducible System Negative Control Repressible System Positive Control Inducible System Positive Control Repressible System Structure of an Operon Inducible Operon Repressible Operon The lac operon • In E. coli, glucose is the preferred carbon source when both glucose and lactose are present. • Jacob and Monod, 1950s, studied lactose metabolism and mutants, and won a Nobel Prize in 1965. Kinetics of induction of lactose operon mRNA and proteins Characteristics of partial diploids containing several combinations of lacI, lacO and lacP alleles Mutations of lac operon Mutation Effect lacIRepressor protein cannot bind, constitutive expression results. lacIs Repressor binds tightly to operator, not inducible. lacOc Repressor cannot bind to the operator site; constitutive expression. (cis-dominant) Mutations of lac operon, cont. Mutation Effect lacPRNA polymerase cannot bind, no transcription results. lacZNo b-galactosidase synthesis. lacYNo permease synthesis. Mutations of lac operon, cont. Mutation Polar mutations crp- Effect Nonsense- termination of transcription Catabolite activator protein cannot bind to crp site, no RNA binding, no transcription The 3 structural genes in the lac operon and the mechanism of their regulation by the lac repressor lac operon model • 2 kinds of genes: structural, regulatory elements. • Polycistronic structural genes, with promoter and operator constitute the lac operon. • Promoter mutants make no lac mRNA. • lacI gene makes a repressor, which binds to the operator. • When operator is ‘repressed’ no transcription occurs. • Inducers bind to repressor, lac mRNA is made. Positive regulation of lactose operon • In presence of glucose, lac operon is ‘off’. How? Structure of cyclic adenosine monophosphate (cAMP) Lac operon is negatively regulated by the lac repressor and positively regulated by the cAMP-CRP complex The 4 critical sequences in the lac operon bound by CRP, RNA polymerase, repressor and the ribosome Structure of the tryptophan (trp) operon showing regulatory elements and the structural genes Binding of tryptophan (the co-repressor) activates an inactive repressor into an active form capable of binding to the trp operator site Structure of the 3’-end of a mRNA terminated at a rho-independent termination site Structure of the leader polypeptide in the trp operon The two tandem tryptophans in the leader peptide act as “stalling sequences” in the absence of tryptophan in the cell Alternative conformations that the trp leader RNA can assume which are important in attenuation Other operons with repeated amino acid sequences that act as “stalling sequence” during attenuation