* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Histone Modifications
Genomic library wikipedia , lookup
Epigenetic clock wikipedia , lookup
Biology and consumer behaviour wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Neocentromere wikipedia , lookup
Extrachromosomal DNA wikipedia , lookup
Point mutation wikipedia , lookup
Minimal genome wikipedia , lookup
Human genome wikipedia , lookup
Oncogenomics wikipedia , lookup
Deoxyribozyme wikipedia , lookup
X-inactivation wikipedia , lookup
Epigenetics of cocaine addiction wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Genome (book) wikipedia , lookup
Non-coding DNA wikipedia , lookup
DNA methylation wikipedia , lookup
Gene expression programming wikipedia , lookup
Ridge (biology) wikipedia , lookup
Genome evolution wikipedia , lookup
Microevolution wikipedia , lookup
Genomic imprinting wikipedia , lookup
History of genetic engineering wikipedia , lookup
Helitron (biology) wikipedia , lookup
Primary transcript wikipedia , lookup
Epitranscriptome wikipedia , lookup
Bisulfite sequencing wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Behavioral epigenetics wikipedia , lookup
Gene expression profiling wikipedia , lookup
Designer baby wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Epigenetics of depression wikipedia , lookup
Epigenetics wikipedia , lookup
Cancer epigenetics wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Epigenetics in stem-cell differentiation wikipedia , lookup
Epigenomics wikipedia , lookup
Histone acetyltransferase wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
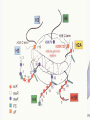
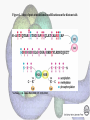
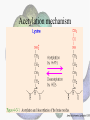
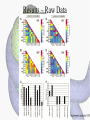
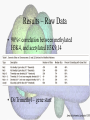
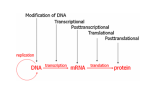
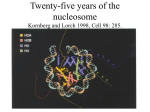
Histone Modifications Seminar in Bioinformatics Saar Gershuni 2005 Background – DNA Packing • The DNA is packed in various levels of condensation in the nucleous (*10,000) • Form of condensation – biological role • Chromosomes, Euchromatin, heterochromatin, DNA strand The Beads on a String • • • • The Histones form the 11nm strand. Are octamer build H3,H4,2HA,2HB monomers 146-147 bp are wrapped around every histone core The histone tail sequences account for 28% of the total amino acid content of the core histones “chromatin fiber folding: requirement for the Histone H4 n-terminal tail “, Benedetta Dorigo†, Thomas Schalch†, Kerstin Bystricky†, ‡ and timothy J. Richmond , journal of molecular biology 327, 1 , 14 march 2003 • Tail of histone h4 taken from cow The Histone Core Wrapped With DNA Chemical Review • Acetyl • Methyl • Phosphoryl • Ubiquitin Histone Modifications • • • • • • De/Acetylation Methylation Phosphorylation Ubiquitination ADP-Rybosilation Swi/Snf complex, which, in vitro, uses the energy of ATP hydrolysis to disrupt histoneDNA interactions Histone Modifications Map Histone Modifications - Role • • • • Transcription – Acetylation/Methylation DNA repair – H2A -Phosphorilation Mitosis – chromosomal arrengement Chromatin assembly – DNA replication Figure 1. Sites of post-translational modificationson the histone tails Yi Zhang et al. Genes Dev. 2001; 15: 2343-2360 Examples of Biological Role of Histone Modifications Acetylation –Lisine • Transcription – loosening the strand • Replication – the positioning of histones • Gcn5 – h3k14 Acetylation mechanism Examples of Biological Role of Histone Modifications Phosphorylation – serine, threonine • Chromosomal condensation – H1,H3 • Rsk-2, an H3 kinase, Coffin Lowry syndrome • Transcription regulation - drosophila sex chromosomes serine 10 H3 in concert with H4K16 (scienceweek) Coffin Lowry Syndrome Coffin-Lowry syndrome is a rare genetic disorder characterized by mental retardation; abnormalities of the head and facial area; large, soft hands with short, thin (tapered) fingers; short stature; and/or various skeletal abnormalities. Characteristic facial features may include an underdeveloped upper jawbone, an abnormally prominent brow, downslanting eyelid folds, widely spaced eyes, large ears, and/or unusually thick eyebrows. Skeletal abnormalities may include abnormal front-to-back and side-to-side curvature of the spine and unusual prominence of the breastbone. Coffin-Lowry syndrome is caused by mutations in the RSK2 gene and is inherited as an X-linked dominant genetic trait. Males are usually more severely affected than females. Examples of Biological Role of Histone Modifications Methylation – Arginine, Lisine – Less studied, enzymology – not known – Can be mono-, bi-, tri- methylated – Transcription regulation - CARM1, argininespecific, histone h3-selective methyltransferase activity, coactivator, with p160 family Figure 2. Chemistry of arginine and lysine methylation Yi Zhang et al. Genes Dev. 2001; 15: 2343-2360 Histone Code • Code - a system of signals or symbols for communication (webster meriam online) • Requirements from a code: – Consistent – Combinatorial (Kurdistany & Grunstein, 2003) Mapping Global Histone Acetylation Patterns to Gene Expression Siavash K. Kurdistani, Saeed Tavazoie, and Michael Grunstein Introduction • The mechanism by which histone de/acetylation affect transcription involve two pathways: – By altering the folding properties of the chromatin fiber – By providing binding surface for recruitment of other elements • To date (6/2004) there is no evidence for consistent patterns of de/acetylation from gene to gene or for the combinatorial use of histone modification sites The Experiment – Data Collection & Methods • Chromatin was extracted from YDS2 exponentially growing • Using chip and DNA microarrays levels of modification was determined • 11 sites of acetylation were examined – H4k8,12,16 H3k9,14,18,23,27 H2ak7,h2bk11,16 The Experiment - Methods • • • • Two Microarrays: 6700 IGR, 6200< ORF 2-4 repetitions Normalization by the ration of total intensities. Coefficient of variation < 0.5 between replicate experiment were counted • End up with 2206 IGR, 2403 ORF • Data were again normalized over the 11 sites per histone ChIP Results – Raw Data Results – Correlation With Gene Expression These results – though significant might be artificially low due to technicalities. Results - Clustering Results – coexpression of Clusters Problem: what is the expression level of randomly selected cluster of genes? The study also checked expression levels at different stress conditions (255) – correlation was found in 6 IGR’s and 13 ORF’s Results – Clusters Biological Relevance • Annotations for all the genes in a cluster were taken from MIPS, GO, MDS • 12/53 IGR’s, 13/68 ORF’s – with significant results • Motif search using AlignACE algorithm found 102 of 29/53 IGR’s, 110 of 34/68 ORF’s Does the Modifications Constitute a Code? • The authors believe that the answer is no because: • The total number of modifications does not contain more information than the sum of individual modification. • Problem: it has been shown to be combinatorial – bdf1 in vitro preference for tetra acetylated H4. Problems • Cutting the chromatin fiber was done using sonicator bath thus creating various size of fiber – with various number of nucleosome – problem with measuring acetylation levels. • Microarrays are 1kb in length can contain up to 5 nucleosomes. Problems • Normalization – step 1 – average number of 1 through all the genome. Step 2 – normalizing groups of 11 lysines in each and every locus (=0, var=1) • All the problems relates with k means algorithm, AlignACE, and gene expression data Genomic Maps and Comparative Analysis of Histone Modifications in Human and Mouse Bradley E. Bernstein, Michael Kamal, Kerstin Lindblad-Toh, Stefan Bekiranov, Dione K. Bailey, Dana J. Huebert, Scott McMahon, Elinor K. Karlsson, Edward J. Kulbokas III, Thomas R. Gingeras, Stuart L. Schreiber, and Eric S. Lander The Experiment • Large scale study of histone modifications (methylation, acetylation) patterns in human and mouse cells • Methods: ChIP, RTPCR – for validating the data, tiling oligonucleotide arrays – 35 bp intervals • Focus: chromosomes 21,22, (H3K4 di/trimethylation and H3K9,14 acetylation) and cytokine cluster, IL4 Receptor, and Hox clusters (H3K4 dimethylation) Results – Raw Data • 90%< correlation between methylated H3K4, and acetylated H3K9,14 • Di/Trimethyl – gene start Conservation of Modification pettern Between Human and Mouse • For this purpose the IL4R methylation analysis were preformed • 55% conservation in human, 68% conservation in mouse, (*7 than random) with no correlation with sequence conservation Hox Clusters • A group of linked regulatory homeobox genes that are involved in patterning the animal body axis during development. Homeobox genes are defined as those that contain an 180-base-pair sequence that encodes a DNA-binding helix–lturn–helix motif (a homeodomain). (Nature) • The remaining orthologous regions between human and mouse Methylation Patterns in Hox Clusters • Completely unique. • Contain huge methylated regions encompass multiple genes • Evolutionary conserved (human-mouse) • Methylation correlates with expression both in ORF’s and IGR’s unlike IL4R Problems • The method of creating the lysate is still sonication. • No relation with genes functionality, cell cycle phase – can change nucleosome concentration What Is It Good for? • Genome wide understanding of chromatin role (structure, functionality( in biology • The use of all the methods we learned • Improve our understanding regarding various mechanisms and processes within the nucleous • Develop new bioinformatic and biological methods for research (advanced ChIP technics, combination with tiling microarrays, and data analyzing tools – normalizations, intergrated tools) Where Do We Go Next? • Characterization of lysine 56 of histone H3 as an acetylation site in Saccharomyces cerevisiae. Ozdemir A, Spicuglia S, Lasonder E, Vermeulen M, Campsteijn C, Stunnenberg HG, Logie C. • Epigenomic mapping in Arabidopsis using tiling microarrays. Martienssen RA, Doerge RW, Colot V. • The epigenetic breakdown of cancer cells: from DNA methylation to histone modifications. Ballestar E, Esteller M. Questions?