* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Document
Human genome wikipedia , lookup
Minimal genome wikipedia , lookup
Molecular cloning wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
DNA vaccination wikipedia , lookup
Epigenetic clock wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Cell-free fetal DNA wikipedia , lookup
Genome evolution wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
DNA methylation wikipedia , lookup
Gene expression programming wikipedia , lookup
DNA supercoil wikipedia , lookup
Oncogenomics wikipedia , lookup
Skewed X-inactivation wikipedia , lookup
Gene expression profiling wikipedia , lookup
Transgenerational epigenetic inheritance wikipedia , lookup
Extrachromosomal DNA wikipedia , lookup
Non-coding DNA wikipedia , lookup
Bisulfite sequencing wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Genome (book) wikipedia , lookup
Genomic imprinting wikipedia , lookup
Epigenetics of depression wikipedia , lookup
Primary transcript wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Point mutation wikipedia , lookup
Designer baby wikipedia , lookup
History of genetic engineering wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Microevolution wikipedia , lookup
Behavioral epigenetics wikipedia , lookup
Helitron (biology) wikipedia , lookup
Neocentromere wikipedia , lookup
Histone acetyltransferase wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
X-inactivation wikipedia , lookup
Epigenetics wikipedia , lookup
Cancer epigenetics wikipedia , lookup
Epigenetics in stem-cell differentiation wikipedia , lookup
Epigenomics wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Epigenetics in learning and memory wikipedia , lookup
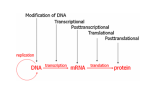
Different types of chromatin heterochromatin Constitutive heterochromatin: euchromatin (and facultative heterochromatin) • constitute ~ 10% of nuclear DNA • highly compacted, transcriptionally inert, replicates late in S phase Euchromatin + facultative heterochromatin: • constitute ~ 90% of nuclear DNA • less condensed, rich in genes, replicates early in S phase however, • only small fraction of euchromatin is transcriptionally active • the rest is transcriptionally inactive/silenced (but can be activated in certain tissues or developmental stages) • these inactive regions are also known as “facultative heterochromatin” Gene silencing and why is it important • In any given cell, only a small percentage of all genes are expressed • vast majority of the genome has to be shut down or silenced • knowing which genes to keep on and which ones to silence is critical for a cell to survive and proliferate normally Gene silencing and why is it important Wolffe and Matzke, Science, 1999 Epigenetics and development n + n Differentiation 2n DNA content same DNA content, > 200 cell types Epigenetics and development De-differentiation? 2n DNA content same DNA content, > 200 cell types examples: 1. Cloning by nuclear transfer --> regenerate entire organism from transfer of single nucleus (e.g. Dolly) 2. Induced pluripotent stem cells (iPS) --> expression of 4 genes are sufficient to transform differentiated cells to “stem” cells Both processes must involve reprogramming of epigenome! Epigenetics and epigenetic regulation Definition of Epigenetics: • heritable changes in gene expression that do not involve changes in DNA sequences • mechanisms: • DNA methylation • histone modifications • examples: • Developmentally regulated or tissue specific gene expression • X chromosome dosage compensation • Drosophila position effect variegation (PEV) Epigenetic mechanism #1: DNA methylation • DNA methylation has long been correlated with repression of gene expression • DNA methylation mostly occurs on CpG dinucleotides DNMTs methyl group is added to the cytosine methylation status is maintained during replication by DNMTs DNA methylation and gene silencing Mechanism of how DNA methylation silences gene expression: • steric hindrance? TF • methylated DNA recruits histone de-acetylases A class of proteins called MBD bind methylated DNA • MeCP2 is the first protein found to bind to methylated DNA • mutation of MeCP2 gene causes Rett Syndrome in humans shifted probes unmethylated probe methylated probe MBD proteins interact with histone deacetylases • MBD2 co-IPs with HDAC activity • MBD2 physically co-IPs with HDACs • MBD2 and HDACs co-purify in the same complex DNA methylation recruits histone deacetylases Epigenetic mechanism #2: histone methylation • histone H3 is methylated at several lysine residues • H3 K4-methylation is associated with transcriptional activation whereas K9-, K27-methylation is associated with repression • these H3 methylation sites define the transcriptional/epigenetic states of the associated genes/chromatin domains Epigenetics example #1: Tissue-specific and developmentally regulated gene expression • globin genes are expressed only in erythroid cells • hemoglobin made up of 2 copies each of a- and b-chains Gene order of globin clusters mirror expression pattern during development HS-40 LCR Globin genes are tissue-specific and developmentally regulated • Distinct isoforms of the globin genes are expressed at different developmental stages • e.g., for the b-globin family, expression goes from e- to g- to b-isoforms • mutations in adult isoforms of globin genes result in thalassemia Globin LCR and adult b-globin promoters are hyperacetylated in adult mouse erythroid leukemia cells upon induction Forsberg et al, PNAS, 2000 Epigenetics example #2 Dosage compensation of X chromosome • for many organisms, females have 2 copies of the X chromosome whereas males only have single copy • how to balance expression dosage of X-linked genes? 2048 identical DNA strands Drosophila polytene chromosomes bands • Drosophila genome has 4 chromosomes • polytene chromosomes result from endoreplication (DNA replication without cytokinesis) inter-bands giant chromosomes that are easily visible X chromosome in Drosophila • the X chromosome of male Drosophila is transcriptionally twice as active • increased transcription of the active X chromosome is marked by hyper-acetylated histones X X DAPI (DNA) Ac H4 X chromosome inactivation • In female mammals, one of the two X chromosomes in the genome is transcriptionally inactivated in order to equalize expression of X-linked genes in males and females (dosage compensation) • Inactivation of the maternal or paternal chromosome is random X chromosome inactivation • In X inactivation, almost the entire X chromosome is transcriptionally silenced • Transcriptional silencing of this chromosome correlates with distinct histone modification patterns • eg. histone H4 is hypo-acetylated on the inactive X chromosome metaphase chromosome immunofluorescence Jeppesen et al, Cell, 1993 The inactive X chromosome is depleted of K4-methylated H3, but is enriched for K27-methylated H3 DAPI DAPI a-MeK4 H3 a-MeK27 H3 MeK4 H3 + DAPI MeK27 H3 + DAPI X inactivation involves sequential epigenetic modifications of the silenced chromosome Epigenetics example #3 Position effect variegation in Drosophila White gene encodes red pigment in eye w+/+ w-/- w+/+ w+/+ mosaic due to PEV Position effect variegation in Drosophila spreading of heterochromatin silencing leads to inactivation of white gene --> mosaic eye patches example of epigenetic regulation since silencing of white gene is NOT due to DNA mutation, but due to translocation and spreading of heterochromatin Position effect variegation in Drosophila Su(var) mutations = Suppressors of PEV e.g. Su(var)2-5 = HP1 Su(var)3-9 = SET-domain protein Identification of H3 Lys9 methyltransferase • The first lysine-specific HMT was identified by IP-in vitro activity assays 1 9 • The SET domain of the SUV39H1 is required for histone methyltransferase activity and this enzyme methylates H3 at Lys9 Rea et al, Nature, 2000 Identification of other H3 methyltransferases • The SET domain is the conserved catalytic core of histone methyltransferases H3: ... ARKSA ... ARTKQTARKSTGGKAPRK 4 9 27 Me Me Me Suv39H1/2 Su(var) 3-9 SET domain human Drosophila Identification of H3 methyltransferases • The SET domain is the conserved catalytic core of histone methyltransferases H3: ... ARKSA ... ARTKQTARKSTGGKAPRK 4 9 27 Me MLL Trx Me Me Suv39H1/2 Su(var) 3-9 EZH2 E(Z) SET domain • Mutations of some histone methyltransferases cancer human Drosophila How does H3 K9-methylation functions in heterochromatin assembly? • back to early genetics studies in Drosophila: • Su(Var) 2-5 (gene) codes for heterochromatin protein 1 (HP1) • HP1 in Drosophila is localized to the chromocenter HP1 DNA Ectopic expression of SUV39H1 causes redistribution of HP1 Melcher et al, MCB, 2000 Lys9-methylated H3 binds to the conserved motif called chromodomain • Using the peptide pull-down assay, it was found that Lys9-methylated H3 binds to heterochromatin protein 1 (HP1) • HP1 is a protein previously identified to be enriched in and important for heterochromatin assembly • Lys9-methylated H3 binds to HP1 via the chromodomain motif in HP1 Bannister et al, Nature, 2001 H3 K9-methylation is required for HP1 localization Lachner et al, Nature, 2001 H3 K9-methylation is required for HP1 localization Lachner et al, Nature, 2001 Histone modification-dependent recruitment of proteins Transcriptional activation TAFII250 Bromodomain Ac H3 ... ARKSTGGK ... 9 14 Histone modification-dependent recruitment of proteins Heterochromatin assembly, Transcriptional silencing Transcriptional activation HP1 TAFII250 Chromodomain Bromodomain Me H3 Ac ... ARKSTGGK ... 9 14 Histone methylation is important for defining and maintaining epigenetic states Identifying methyl-H3 binding proteins • histone peptide pulldown assay: b a ? b b a ? a = candidate approach b = unbiased approach identify by Western blotting identify by Mass Spec Site specific methylation of the H3 tail has different functions BPTF PhD Me H3: HP1 polycomb CD CD Me Me ... ARKSA ... ARTKQTARKSTGGKAPRK 4 9 27 transcriptional “competence” transcription repression transcription repression euchromatin constitutive heterochromatin facultative heterochromatin Heterochromatin and euchromatin constitutive heterochromatin K9-methylated H3 HP1 facultative heterochromatin K27-methylated H3 polycomb euchromatin K4-methylated H3 BPTF Yng2 Different dynamics of histone modifications HATs Ac-histone histone HDACs highly dynamic kinases Phos-histone histone phosphatases HMT Me-histone histone de-methylase more stable The search for histone demethylases • LSD1 is a transcriptional co-repressor and its repression function is mediate through the amine-oxidase domain Transcription ? luciferase 5X Gal4 binding sites Shi et al, Cell, 2004 The search for histone demethylases • LSD1 is a histone H3-K4 demethylase Shi et al, Cell, 2004 The search for histone demethylases • LSD1 is a histone H3-K4 demethylase Shi et al, Cell, 2004 The search for histone demethylases Adapted from Tsukada and Zhang, Methods, 2006 Purifcation of histone demethylases Release of radioactive formaldehyde Adapted from Tsukada and Zhang, Methods, 2006 Identifying site of histone demethylation • JHDM1A demethylates di-MeK36 on H3 Adapted from Tsukada and Zhang, Methods, 2006 Overexpression of JHDM1A results in loss of K36MeH3 Adapted from Tsukada and Zhang, Methods, 2006 Histone de-methylases are found for all these sites: LSD1 JARID1a-d JMJD2b UTX JMJD3 Apart from LSD1, all other histone de-methylases identified so far belong to the JmjC domain-containing family of enzymes Epienetics and diseases diseases • b-globin thalassemia • leukemia adapted from Nature 429, 2004 Paper assignments for Nov 2nd: Group 1 (Chanda - Fasih): Kuzmichev et al, 2002, Genes Dev. 16: 2893-2905 Histone methyltransferase activity associated with a human multiprotein complex containing the enhancer of Zeste protein Group 2 (Fenton - How): de Napoles et al, 2004, Dev. Cell 7: 663-676 Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation Group 3 (Karisch - Yan): Wysocka et al, 2006, Nature 442: 86-90 A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodeling