* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download In situ - University of Evansville Faculty Web sites
Human genetic variation wikipedia , lookup
Cancer epigenetics wikipedia , lookup
Copy-number variation wikipedia , lookup
Oncogenomics wikipedia , lookup
Genome (book) wikipedia , lookup
Gel electrophoresis of nucleic acids wikipedia , lookup
United Kingdom National DNA Database wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
DNA barcoding wikipedia , lookup
Mitochondrial DNA wikipedia , lookup
Genetic engineering wikipedia , lookup
DNA vaccination wikipedia , lookup
Minimal genome wikipedia , lookup
Comparative genomic hybridization wikipedia , lookup
Nucleic acid double helix wikipedia , lookup
Zinc finger nuclease wikipedia , lookup
DNA sequencing wikipedia , lookup
SNP genotyping wikipedia , lookup
Genealogical DNA test wikipedia , lookup
DNA supercoil wikipedia , lookup
Designer baby wikipedia , lookup
Primary transcript wikipedia , lookup
Transposable element wikipedia , lookup
Molecular cloning wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Point mutation wikipedia , lookup
Epigenomics wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Extrachromosomal DNA wikipedia , lookup
Public health genomics wikipedia , lookup
Microevolution wikipedia , lookup
Pathogenomics wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Cell-free fetal DNA wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
History of genetic engineering wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Bisulfite sequencing wikipedia , lookup
Whole genome sequencing wikipedia , lookup
Human genome wikipedia , lookup
Human Genome Project wikipedia , lookup
Genome evolution wikipedia , lookup
Non-coding DNA wikipedia , lookup
Metagenomics wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Microsatellite wikipedia , lookup
Genome editing wikipedia , lookup
Helitron (biology) wikipedia , lookup
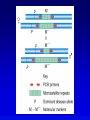
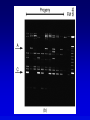
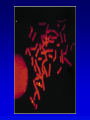
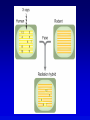
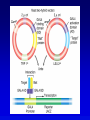
Chapter 9 Genomics Mapping and characterizing whole genomes 16 and 20 February, 2004 Overview • Genomics is the molecular mapping and characterization of whole genomes and whole sets of gene products. • Consecutive high-resolution genetic and physical maps culminate in the complete DNA sequence. • Sequencing strategies depend upon the size of the genome and the distribution of its repetitive sequences. • Assembly of sequences is done clone by clone or by whole genome assembly, or both. • Computational analysis is used to describe encoded information whereas functional genomics explores function and interaction of gene products. Genomics • Focuses on the entire genome • Made possible by advances in technology – automated cloning and sequencing (robotics) allowing high throughput – computerized tracking and analysis of sequences • Insights into global organization, expression, regulation and evolution – enumeration of genes – identification regulatory and functional motifs • Functional genomics to determine actual function of genetic material Genome projects • Starts with high-resolution recombination and cytogenetic maps of each chromosome • Followed by physical characterization and positioning of cloned DNA fragments to anchor to high-resolution map • Followed by large-scale sequencing and analysis – clone-based sequencing – whole genome shotgun sequencing • Last step: functional genomics (the hard part) High-resolution genetic maps •Start with low-resolution maps from existing recombination maps •Next, layer DNA polymorphisms onto map –e.g., neutral DNA sequence variation not associated with phenotypic variation •phenotypic consequences, if any, irrelevant –such DNA markers behave as allelic gene pairs and can be detected by Southern blotting or PCR –mapped by recombination or cytogenetics RFLP • • • • Restriction fragment length polymorphism May or may not be neutral Detected by Southern blotting or PCR Multiple RFLPs can be mapped by classical segregation analysis • Example: A a | = cut site = primer – restriction digestion of PCR products yields one fragment for allele A and two fragments for a – note that homozygotes and heterozygote have different restriction patterns, permitting identification of carrier Physical maps • Maps of physically isolated pieces of genome, i.e., cloned DNA – previously cloned DNA can be localized to map by Southern blotting or PCR to measure location and distance – useful in assembly of sequences • Vectors with large inserts are most useful • Overlapping clones are assembled into contigs, ideally, one per chromosome – mapping of restriction sites – sequence-tagged sites (STSs) Short-sequence repeat markers • Tandemly repeated • Variable numbers of repeats, give different size restriction fragments detected on Southern blots • Single sequence length polymorphisms (SSLPs) – – – – e.g., TGACGTATGACGTATGACGTATGACGTA mutations give rise to large number of alleles higher proportion of heterozygotes two types in genomics • minisatellite (VNTRs) • microsatellite Minisatellites and microsatellites • Minisatellites – based on variation of number of tandem repeats (VNTRs) which segregate as alleles – in humans, repeat unit is 15-100 nucleotides, for total of 1-5 kb – if number of repeats is variable, Southern blot will show numerous bands – basis of DNA fingerprinting and can be used in mapping • Microsatellites – sequences dispersed throughout the genome – variable numbers of dinucleotide repeats – detected by PCR RAPDs • Randomly amplified polymorphic DNA • PCR primers with random sequences often amplify one or more regions of DNA – primer complement randomly located in genome – single primer can detect regions with inverted repeats – polymorphisms segregate as alleles and therefore can be mapped in crosses • Often used in evolutionary studies Human high-resolution map •RFLP, SSLP, and RAPD markers have been mapped to 1cM density •Provide landmarks for anchoring sequence information •1 cM of human DNA is ~ 1 Mb of DNA, still a large amount •Single nucleotide polymorphisms (SNPs) estimated number about 3 million between any two genomes High-resolution cytogenetic maps • Relates markers to chromosome bands, puffs or disruptions • In situ hybridization – cloned DNA labeled with radioactivity or fluorescent dye (FISH) – hybridized to denatured metaphase or polytene chromosomes – indicates approximate locations – FISH extension allows chromosome painting • Rearrangement breakpoint mapping – detected by Southern blotting Assembling genomes with repetitive sequences • Use of ordered clones – e.g., C. elegans – large mapped, cosmids with minimum overlap (minimum tiling path) subcloned into sequencing vectors – inserts sequenced by automated methods – sequence assembled by computer based on map • Whole genome shotgun – e.g., D. melanogaster – three libraries (2-kb, 100-kb, 150-kb) of genomic clones, each sequenced from both ends – sequences aligned by homologous sequence overlap and by use of paired-end sequences to produce scaffolds of contigs Assembling genomes • If genome is rich in repetitive elements, contigs may be short • Gaps usually occur, regardless of technique – short gaps filled by PCR – long gaps require additional cloning, sometimes in different host • Sequenced eukaryotic genomes include: Saccharomyces cerevisiae, Caenorhabditis elegans, Drosophila. melanogaster, Arabidopsis thaliania, Mus musculus, Danio rerio, Homo sapiens Bioinformatics (1) • Not clear what all of nucleotide sequence of draft genome means • In addition to proteome (protein encoding sequences), genome contains additional information • Considerable ignorance due to the following: – docking (target) sequences of many DNA binding proteins are unknown – alternative splicing complicates ORF finding – some sequences have context-dependent meaning – some sequences have multiple uses • Bioinformatics and functional genomics attempt to decipher genome Bioinformatics (2) • Uses available information (much of it available on the Web) to predict function of sequences • cDNA evidence – motifs, e.g., start codons, ORFs – expressed sequence tags (ESTs), from reverse transcribed mRNA • mRNA and ORF structure – gene and intron finding programs • Polypeptide similarity evidence – at level of >35% sequence identity, polypeptides likely have common function – often identified by BLASTp search • Codon bias information Functional genomics • Study of expression and interaction of gene products • Requires new vocabulary and techniques – transcriptome: all DNA transcripts • may be monitored by use of DNA chips – proteome: all encoded proteins • complicated by alternative splicing – interactome: all interactions between all categories of molecules • detected by two-hybrid system and related procedures – phenome: phenotype of each gene knockout Assignment: Continue with * section of the Web tutorial.