* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Cell Assemblies - CAAM @ Rice
Molecular neuroscience wikipedia , lookup
Neuromuscular junction wikipedia , lookup
Convolutional neural network wikipedia , lookup
Metastability in the brain wikipedia , lookup
Development of the nervous system wikipedia , lookup
Neural coding wikipedia , lookup
Central pattern generator wikipedia , lookup
Clinical neurochemistry wikipedia , lookup
Premovement neuronal activity wikipedia , lookup
Mirror neuron wikipedia , lookup
Types of artificial neural networks wikipedia , lookup
Neurotransmitter wikipedia , lookup
Multielectrode array wikipedia , lookup
Neuroanatomy wikipedia , lookup
Optogenetics wikipedia , lookup
Nonsynaptic plasticity wikipedia , lookup
Chemical synapse wikipedia , lookup
Pre-Bötzinger complex wikipedia , lookup
Feature detection (nervous system) wikipedia , lookup
Single-unit recording wikipedia , lookup
Stimulus (physiology) wikipedia , lookup
Electrophysiology wikipedia , lookup
Biological neuron model wikipedia , lookup
Channelrhodopsin wikipedia , lookup
Neuropsychopharmacology wikipedia , lookup
Synaptic gating wikipedia , lookup
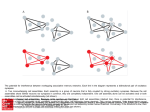
Understanding a Cell Assembly Down to its Core Karina Aliaga, Department of Mathematical Sciences, New Jersey Institute of Technology Tyler Young, Department of Bioengineering, Rice University Dr. Steve Cox, Department of Computational and Applied Mathematics, Rice University Dr. Illya Hicks, Department of Computational and Applied Mathematics, Rice University Cell Assemblies and Graph Theory Introduction Ever since psychologists and neuroscientists began studying the physiological inner workings of the brain, they have been puzzled by many questions. How are concepts stored and recalled within our brains? How does learning and memory occur? In 1949, D.O. Hebb tried to explain the answers to these questions in terms of cell assemblies in his book The Organization of Behavior. Hebb asserts that a cell assembly is a group of neurons wired in a specific manner such that when a sufficient amount of neurons in this group are excited, the entire group becomes excited in a synchronized manner. Hebb went on to explain that these cell assemblies form via synaptic plasticity. He claims that if neuron A repeatedly fires neuron B, some metabolic activity occurs increasing the efficiency in which neuron A fires neuron B making it easier for neuron A to fire neuron B. This phenomenon is more commonly known as “cells that fire together, wire together.” Hebb postulates that the ignition of a series of these groups of neurons, or cell assemblies, can explain how concepts are stored and recalled within our brains, thus allowing learning and memory to occur. -cores: a concept that many graph theorist have been studying since the 1970’s. A -core can be defined as: Given graph X, the subset X is a _-core if every node in _ has at least _ neighbors in X _ . Minimal _-core: If no proper subset of the _-core is also a _-core, then it is minimal. Examples 2 Closure: 1 3 (1) Create : we will use the minimal 2-core found by previous example 5 4 2 3 1 4 6 Cell Assemblies and k-cores (2) Find in the 5 : One type of cell assembly, we will call a -assembly, is the closure of a minimal core. In order to find these -assemblies, we needed to develop a method for finding minimal -cores and their closures in any given network of neurons. Research Goal Algorithms Our research goal is to understand cell assemblies and to develop a method for finding them within any given network of neurons. This will allow us to better understand and explain the inner workings of our brains. Finding minimal _-cores: MATLAB has a built-in function (________) that finds an optimal solution to binary integer programming problems of the following form: Because we must apply again: minimizes the objective function: ________ arguments: : Coefficients of the variables of the objective function : Using the threshold inequality we can find _ _: Applying again will generate an invariant set because the whole graph is excited, thus giving us the closure of , a - assembly. Cell Assemblies: A Different Perspective G. Palm: Towards a Theory of Cell Assemblies. Biological Cybernetics.181-193 (1981), was the first to give a mathematical definition of cell assemblies. Let be a graph with a set of vertices Adjacency Matrix ( and edges ) A matrix of binary elements representing the connectivity of a network of neurons such that if_______ __ there exists a connection between neurons and _ . Conversely, if ___then no connection between neurons _ and _ exists. Finding _-assemblies: ______ minimizes __ constrained to ______. Because satisfies the inequality, we must add an additional constraint. _ Threshold ( ) The minimum number of inputs a neuron needs in order to become excited Closure :The invariant set generated by Let be a subgraph in . . Conclusion Examples is a binary vector representing the presence ( ) or absence ( ) of a neuron 2 3 1 4 6 A set is invariant if: 5 In this example, finds the minimal 2-core . There are, however, many other minimal 2-cores in this set of neurons such as , and ` . Our goal for the future is to find some method that enables ______ to find all of the minimal -cores in a given network thus, allowing us to find all of the assemblies in that network. Working on this project, we have extended Palm’s Mathematical definition of cell assemblies to fit a binary integer programming problem. Also, we created a link between the world of graph theory and cell assemblies: the closure of a minimal _-core is a - assembly. Lastly, we created an algorithm that allows us to find at least one cell assembly in any given network of neurons. Acknowledgements This work was supported by a NSF REU Grant DMS-0755294 and NSF VIGRE Grant DMS-0240058. We would like to thank Dr. Cox and Dr. Hicks for their guidance throughout this project. Lastly, we would like to thank Diane Taylor and Shaunak Das, our research collaborators.