* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Membrane targeting of proteins
Cell nucleus wikipedia , lookup
Lipid bilayer wikipedia , lookup
Protein phosphorylation wikipedia , lookup
Magnesium transporter wikipedia , lookup
Theories of general anaesthetic action wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Bacterial microcompartment wikipedia , lookup
Cell membrane wikipedia , lookup
SNARE (protein) wikipedia , lookup
Model lipid bilayer wikipedia , lookup
Protein moonlighting wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
List of types of proteins wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Intrinsically disordered proteins wikipedia , lookup
Signal transduction wikipedia , lookup
Endomembrane system wikipedia , lookup
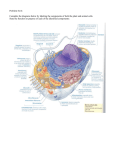
Chapter 3 Membrane targeting of proteins By D. Thomas Rutkowski & Vishwanath R. Lingappa 3.1 Introduction • Cells must localize proteins to specific organelles and membranes. • Proteins are imported from the cytosol directly into several types of organelles. 3.1 Introduction • The endoplasmic reticulum (ER): – is the entry point for proteins into the secretory pathway – is highly specialized for that purpose • Several other organelles and the plasma membrane receive their proteins by way of the secretory pathway. 3.2 Proteins enter the secretory pathway by translocation across the ER membrane (an overview) • Signal sequences target nascent secretory and membrane proteins to the ER for translocation. • Proteins cross the ER membrane through an aqueous channel that is gated. 3.2 Proteins enter the secretory pathway by translocation across the ER membrane • Secretory proteins translocate completely across the ER membrane; – transmembrane proteins are integrated into the membrane. • Before leaving the ER, proteins are modified and folded by enzymes and chaperones in the lumen. 3.3 Proteins use signal sequences to target to the ER for translocation • A protein targets to the ER via a signal sequence, a short stretch of amino acids that is usually at its amino terminus. • The only feature common to all signal sequences is a central, hydrophobic core that is usually sufficient to translocate any associated protein. 3.4 Signal sequences are recognized by the signal recognition particle (SRP) • SRP binds to signal sequences. • Binding of SRP to the signal sequence slows translation so that the nascent protein is delivered to the ER still largely unsynthesized and unfolded. 3.4 Signal sequences are recognized by the signal recognition particle (SRP) • The structural flexibility of the M domain of SRP54 allows SRP to recognize diverse signal sequences. 3.5 An interaction between SRP and its receptor allows proteins to dock at the ER membrane • Docking of SRP with its receptor brings the ribosome and nascent chain into proximity with the translocon. • Docking requires the GTP binding and hydrolysis activities of SRP and its receptor. 3.6 The translocon is an aqueous channel that conducts proteins • Proteins translocate through an aqueous channel composed of the Sec61 complex, located within the ER membrane. • Numerous accessory proteins that are involved in: – Translocation – Folding – Modification associate with the channel 3.7 Translation is coupled to translocation for most eukaryotic secretory and transmembrane proteins • An interaction between the translocon and the signal sequence causes the channel to open and initiates translocation. • The exact mechanism of translocation may vary from one protein to another. 3.8 Some proteins target and translocate posttranslationally • Posttranslational translocation proceeds independently of both ribosomes and SRP. • Posttranslational translocation is used extensively in yeast but is less common in higher eukaryotes. 3.8 Some proteins target and translocate posttranslationally • The posttranslational translocon is distinct in composition from the cotranslational translocon, but they share the same channel. 3.9 ATP hydrolysis drives translocation • The energy for posttranslational translocation comes from ATP hydrolysis by the BiP protein within the ER lumen. • The energy source for cotranslational translocation is less clear, but might be the same as for posttranslational translocation. 3.9 ATP hydrolysis drives translocation • Most translocation in bacteria occurs posttranslationally through a channel that is evolutionarily related to the Sec61 complex. 3.10 Transmembrane proteins move out of the translocation channel and into the lipid bilayer • The synthesis of transmembrane proteins requires that transmembrane domains be – recognized – integrated into the lipid bilayer 3.10 Transmembrane proteins move out of the translocation channel and into the lipid bilayer • Transmembrane domains exit the translocon by moving laterally through a protein-lipid interface. 3.11 The orientation of transmembrane proteins is determined as they are integrated into the membrane • Transmembrane domains must be oriented with respect to the membrane. • The mechanism of transmembrane domain integration may vary considerably from one protein to another – especially for proteins that span the membrane more than once 3.12 Signal sequences are removed by signal peptidase • Nascent chains are often subjected to covalent modification in the ER lumen as they translocate. • The signal peptidase complex cleaves signal sequences. 3.13 The lipid GPI is added to some translocated proteins • GPI addition covalently tethers the C-termini of some proteins to the lipid bilayer. 3.14 Sugars are added to many translocating proteins • Oligosaccharyltransferase catalyzes N-linked glycosylation on many proteins as they are translocated into the ER. 3.15 Chaperones assist folding of newly translocated proteins • Molecular chaperones associate with proteins in the lumen and assist their folding. 3.16 Protein disulfide isomerase ensures the formation of the correct disulfide bonds as proteins fold • Protein disulfide isomerases catalyze disulfide bond formation and rearrangement in the ER. 3.17 The calnexin/calreticulin chaperoning system recognizes carbohydrate modifications • Calnexin and calreticulin escort glycoproteins through repeated cycles of chaperoning. – The cycles are controlled by addition and removal of glucose. 3.18 The assembly of proteins into complexes is monitored • Subunits that have not yet assembled into complexes are retained in the ER by interaction with chaperones. 3.19 Terminally misfolded proteins in the ER are returned to the cytosol for degradation • Translocated proteins can be exported to the cytosol. • There they are: – ubiquitinated – degraded by the proteasome —a process known as ER-associated degradation. 3.19 Terminally misfolded proteins in the ER are returned to the cytosol for degradation • Proteins are returned to the cytosol by the process of retrograde translocation. – This is not as well understood as for translocation into the ER. 3.20 Communication between the ER and nucleus prevents the accumulation of unfolded proteins in the lumen • The unfolded protein response: – monitors folding conditions in the ER lumen – initiates a signaling pathway that increases the expression of genes for ER chaperones • The protein Ire1p mediates the unfolded protein response in yeast by becoming activated in response to conditions of cellular stress. 3.20 Communication between the ER and nucleus prevents the accumulation of unfolded proteins in the lumen • Activated Ire1p splices HAC1 mRNA. • It results in the production of the Hac1 protein, a transcription factor that: – localizes to the nucleus – binds to the promoters of genes with a UPR response element • The unfolded protein response in higher eukaryotes has evolved more layers of control beyond those seen in yeast. 3.21 The ER synthesizes the major cellular phospholipids • The major cellular phospholipids are synthesized predominantly on the cytosolic face of the ER membrane. 3.21 The ER synthesizes the major cellular phospholipids • The localization of enzymes involved in lipid biosynthesis can be controlled by the cell to regulate the generation of new lipids. • Cholesterol biosynthesis is regulated by proteolysis of a transcription factor integrated into the ER membrane. 3.22 Lipids must be moved from the ER to the membranes of other organelles • Each organelle has a unique composition of lipids. – This requires that lipid transport from the ER to each organelle be a specific process. • The mechanisms of lipid transport between organelles are unclear. – They might involve direct contact between the ER and other membranes in the cell. • Transbilayer movement of lipids establishes asymmetry of membrane leaflets. 3.23 The two leaflets of a membrane often differ in lipid composition • Movement of lipid molecules between the leaflets of a bilayer is required to establish asymmetry. • Enzymes (“flippases”) are required for movement of lipids between leaflets. 3.24 The ER is morphologically and functionally subdivided • The ER is morphologically subdivided into specialized compartments, including: – the rough ER for protein secretion – the smooth ER for steroidogenesis and drug detoxification – the sarcoplasmic reticulum for calcium storage and release 3.24 The ER is morphologically and functionally subdivided • The functions of the smooth ER can be specialized according to the needs of the particular cell type. • The ER may also be subdivided at the molecular level, in ways not morphologically evident. 3.25 The ER is a dynamic organelle • The extent and composition of the ER change in response to cellular need. • The ER moves along the cytoskeleton. 3.25 The ER is a dynamic organelle • The mechanisms by which the ER expands and contracts and forms tubules have yet to be discovered. • The signaling pathways that control ER composition are not yet understood but may overlap with the unfolded protein response. 3.26 Signal sequences are also used to target proteins to other organelles • Signal sequences are used for targeting to and translocation across the membranes of other organelles. • Mitochondria and chloroplasts are enclosed by a double membrane, with each bilayer containing its own type of translocon. • Two distinct pathways target matrix proteins to peroxisomes. 3.27 Import into mitochondria begins with signal sequence recognition at the outer membrane • Mitochondria have an inner and an outer membrane, each of which has a translocation complex. • Import into mitochondria is posttranslational. 3.27 Import into mitochondria begins with signal sequence recognition at the outer membrane • Mitochondrial signal sequences are recognized by a receptor at the outer membrane. 3.28 Complexes in the inner and outer membranes cooperate in mitochondrial protein import • The TOM and TIM complexes associate physically, and the protein being imported passes directly from one to the other. • Hsp70 in the mitochondrial matrix and the membrane potential across the inner membrane provide the energy for import. 3.29 Proteins imported into chloroplasts must also cross two membranes • Import into chloroplasts occurs posttranslationally. • The inner and outer membranes have separate translocation complexes that cooperate during the import of proteins. 3.30 Proteins fold before they are imported into peroxisomes • Peroxisomal signal sequences are: – recognized in the cytosol – targeted to a translocation channel • Peroxisomal proteins are imported after they are folded. 3.30 Proteins fold before they are imported into peroxisomes • The proteins that recognize peroxisomal signal sequences remain bound during import and cycle in and out of the organelle. • Peroxisomal membranes originate by budding from the ER.