* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Developing a CRISPR/Cas9 System for Volvox Carteri
RNA interference wikipedia , lookup
Genome (book) wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Epitranscriptome wikipedia , lookup
Quantitative trait locus wikipedia , lookup
Public health genomics wikipedia , lookup
Cancer epigenetics wikipedia , lookup
Ridge (biology) wikipedia , lookup
History of RNA biology wikipedia , lookup
Designer baby wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Oncogenomics wikipedia , lookup
Pathogenomics wikipedia , lookup
Short interspersed nuclear elements (SINEs) wikipedia , lookup
Transposable element wikipedia , lookup
Metagenomics wikipedia , lookup
Point mutation wikipedia , lookup
Microevolution wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Primary transcript wikipedia , lookup
RNA silencing wikipedia , lookup
Minimal genome wikipedia , lookup
History of genetic engineering wikipedia , lookup
Genomic imprinting wikipedia , lookup
Human genome wikipedia , lookup
Helitron (biology) wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Non-coding RNA wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Gene expression programming wikipedia , lookup
Non-coding DNA wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Gene expression profiling wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Genomic library wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Genome evolution wikipedia , lookup
Genome editing wikipedia , lookup
Mir-92 microRNA precursor family wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
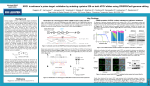
Developing a CRISPR/Cas9 System for Volvox Carteri Robyn Jasper, Jose Ortega, Stephen Miller, PhD Department of Biological Sciences, University of Maryland, Baltimore County Methods Abstract Genome editing is a tool implemented to test gene function through targeted mutations. The Cas9/CRISPR system is simpler and more precise than previously developed genome editing systems. The high precision is due to the CRISPR associated (Cas) endonuclease’s ability to bind DNA via associated guide RNAs. Cas endonucleases can delete or add bases to the genome, which permits not only knockouts to determine mutant phenotypes, but also tagging genes with reporters. However, a Cas9/CRISPR system has not been adapted for use with green algae. In this project, I adapted an existing Cas9 vector for use in the alga Volvox using molecular cloning techniques to insert species-specific regulatory sequences and guide RNA sequence targeting a test gene with known mutant phenotype. Biolistic transformation of the vector resulted in viable transformants, which were tested for guide RNA expression and Cas9 protein expression via RT-PCR and Western blots, respectively. RT-PCR confirmed that guide RNA was made. Cas9 expression is being tested currently. Once transformants that express both components are obtained, we will characterize them for mutations. Ultimately, this system will be used to edit genes related to multicellularity in Volvox and to improve the alga Chlorella as a biofuels and neutraceuticals production organism. Both sgRNA and Cas9 are expressed, but the transformants do not exhibit mutant phenotype Conclusion • Transformants contain cas9 sequence • Cas9 is being expressed when induced • Cas9 accumulation is not toxic to the cells • sgRNA is being expressed Functionality testing: • Vector integration into genome via gDNA PCR • Cas9 expression via Western Blot • sgRNA expression via RT PCR • Identify mutant phenotype via microscopy • The expression cassette is viable for driving expression • No notable mutant phenotype Is the Cas9 active? Is the sgRNA folding properly and active? Cas9 Expressed in Transformants Future Directions Volvocine algae and Volvox carteri induced repressed Kirk 2003 • Common unicellular ancestor ~200 MYA • Multicellular • Varying from unicellular to multicellular, and varying divisions of labor • Full cell differentiation • Easily cultured in lab • Mutants easily isolated • New ATG start codon • RBCS2 intron for increased expression of Cas9 • New sgRNA targets • Cas9 not toxic to transformants • Cas9 expression induced and repressed • Is it functional? • Transform new plasmids and isolate more transformants • Test Cas9 functionality by in vitro assays and immunolocalization sgRNA Expressed in Transformants CRISPR/Cas9 CRISPR - Clustered Regularly Interspaced Short Palindromic Repeats • Test sgRNA accumulation and identify the transcription start site A functioning system can be adapted to: • investigate multicellularity in Volvox • investigate orthologs in Chlamydomonas • investigate metabolic pathways for biofuel production Bacterial genomic sequence involved in immune defense Consists of repeat sequences and foreign DNA spacers Acknowledgements Mostly used to: -Create knockdowns -Insert reporter genes Hsu 2014 U6 regulatory sequences are able to drive sgRNA expression Meyerhoff Scholars Program HHMI Scholars Program MARC U*STAR URA Award