* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Lec206
DNA vaccination wikipedia , lookup
Zinc finger nuclease wikipedia , lookup
Neuronal ceroid lipofuscinosis wikipedia , lookup
Saethre–Chotzen syndrome wikipedia , lookup
Molecular cloning wikipedia , lookup
Genomic imprinting wikipedia , lookup
Long non-coding RNA wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Oncogenomics wikipedia , lookup
Gene nomenclature wikipedia , lookup
Genome evolution wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
X-inactivation wikipedia , lookup
Gene desert wikipedia , lookup
RNA interference wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Genetic engineering wikipedia , lookup
Epigenetics in learning and memory wikipedia , lookup
Non-coding RNA wikipedia , lookup
Epigenetics in stem-cell differentiation wikipedia , lookup
Genome (book) wikipedia , lookup
Primary transcript wikipedia , lookup
Genomic library wikipedia , lookup
Helitron (biology) wikipedia , lookup
RNA silencing wikipedia , lookup
Gene expression profiling wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Gene therapy wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Point mutation wikipedia , lookup
Gene expression programming wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Genome editing wikipedia , lookup
History of genetic engineering wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Mir-92 microRNA precursor family wikipedia , lookup
Designer baby wikipedia , lookup
Microevolution wikipedia , lookup
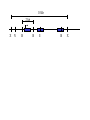
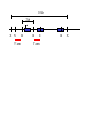
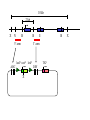
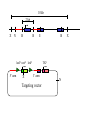
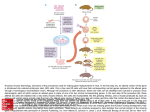
Next lecture:techniques used to study the role of genes in develpoment • • • • • • Random genetics followed by screening Targeted mutagenesis (gene knockout) Transgenic animal models Dominant negative mutant molecules Antisense RNA interference RNA interference (c. elegans-website 4.8) Random genetics • • • • • Chemical mutagen/gene disrupting agent Screening for a phenotype Dominant mutations can be found easily Recessive mutations require breeding Frequently used with model organisms – Especially Drosophila (lectures in late Feb.) • Becoming newly popular in the mouse Mouse ENU mutagenesis Recent applications of this technology can be seen in: Nature Genetics,(Aug 2000)Volume 25 pp.440-443 and 444-447 Trans-heterozygous phenotypes • When two genes are in the same “pathway” mutants heterozygous for both genes will display a phenotype even though each individual heterozygous mutant does not • Can be combined with ENU mutagenesis to screen for genes in the same pathway as another known “knocked out” gene. Targeted mutagenesis (knockout) • Determine the action of a known, cloned gene in a developmental process • Removes a segment of the known gene by homologous recombination • Required elements: – Mapped genomic clone for the gene of interest – Embryonic stem (ES) cells – A lot of repetitive work Points to make • Genomic clone should come from the same mouse strain from which the ES cell is derived (common strain 129SV) • Making of targeting construct. 6kb of homologous arms with appropriately arranged selectable markers for positive and negative selection • A screening strategy involving two probes 10 kb 2 kb X N B B E B X 10 kb 2 kb X N 5’ arm B B E 3’ arm B X 10 kb 2 kb X N B 5’ arm ABC B E B 3’ arm loxP neoR loxP X DEF TKS X 10 kb 2 kb X N B B E loxP neoR loxP 5’ arm X B TKS 3’ arm Targeting vector N X 10 kb 2 kb X N B B E B TKS 5’ arm X 3’ arm Targeting vector N X 10 kb 2 kb X N B B E B TKS 5’ arm X 3’ arm Targeting vector N X 10 kb 2 kb X N E X Homologous recombinant X Targeting vector Randomly integrated B X 10 kb 2 kb X N E X B X Homologous recombinant TKS (gancyclovir) X Targeting vector Randomly integrated 10 kb 2 kb X N 5’ probe E X Homologous recombinant B X 3’ probe X Targeting vector Randomly integrated Note, probes are OUTSIDE the homology arms. Therefore, they will only detect the endogenous locus and the recombinant Removing the selection marker • Cre recombinase deletes sequences between two lox-P sites in the same orientation • Transiently transfect a vector expressing the cre recombinase for deletion • Grow clones again and screen for the deletion by southern blot as before • Especially necessary in studying knockouts of genetic regulatory sequences Cre-mediated deletion 10 kb 2 kb X N X 5’ probe E B Homologous recombinant X 3’ probe Cre expression vector X N 5’ probe E Cre-deleted recombinant B X 3’ probe The next steps • Grow up clones with correct recombinant • Inject ES cells into mouse blastocyst (d3.5) • The ES cells will integrate themselves into the blastocyst and mouse will be a chimera • Usually the ES cell strain has a different coat color than the blastocyst strain so that the “marbleized” mice can be easily seen And then…. See also fig 4.19 on page 98 of Gilbert • Breed the chimeric mice to normal mice • If the ES cells contributed to the germline the babies which contain the mutation will have the coat color of the ES cell – These mice are only HETEROZYGOUS for the targeted gene • These mice need to be bred to homozygous • Analyze the phenotype…... Further references on homologous recombination in ES cells • Capecchi, MR (1989) Altering the genome by homologous recombination. Science. 244:1288-1292. • Ramirez-Solis, R, Davis, AC and Bradley, A. (1993) Gene targeting in embryonic stem cells. Meth. Enzymol. 225:855-875. Possible phenotypes • Something related to what you expected • Something completely unexpected – No phenotype – Embryonic lethal – Complex phenotype-multiple tissues and effects Conditional gene targeting • Tissue-specific knockout of a gene – Avoids embryo lethality – Avoids complex phenotypes • Inducible knockout – Allows “before” and “after” type analysis – Model of acquired mutation rather than inherited mutation Strategy (from Rajewsky, et. al.) Transgenic animals (mice) Points to make • • • • • • • Transgene should be free of vector DNA Transgene must be rigorously purified Transgene integration is a bit inefficient Founder mice are sometimes mosaic Need to outcross the mice to wild type mice The transgene is not always expressed Position-effect-variegation Transgenesis and developmental studies • Inappropriate or overexpression of a gene • Dominant negative mutant gene expression • Reporter gene expression for lineage tracing – Fluorescent proteins (GFP, YFP) – Beta-galactosidase (X-gal staining) • Transcriptional regulatory elements – Assuming a position-independent system Antisense and RNA interference • Overexpression of anti-sense RNA – Not the method of choice though it has worked in some instances. • RNA interference in c. elegans – Website 4.8 Model of RNA interference RNA interference in C. elegans Next lectures: Differential Gene expression • Chapter 5 and websites on syllabus • Epigenetic control mechanisms – Histone modification – DNA methylation – Nucleosome disruption “machines” • Promoters and enhancers – Old and new models of enhancer function • Novel transcriptional control sequences