* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download protein synthesis
Magnesium transporter wikipedia , lookup
Ancestral sequence reconstruction wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Protein moonlighting wikipedia , lookup
Peptide synthesis wikipedia , lookup
Molecular evolution wikipedia , lookup
Ribosomally synthesized and post-translationally modified peptides wikipedia , lookup
Cell-penetrating peptide wikipedia , lookup
Protein (nutrient) wikipedia , lookup
Eukaryotic transcription wikipedia , lookup
Western blot wikipedia , lookup
RNA polymerase II holoenzyme wikipedia , lookup
List of types of proteins wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Amino acid synthesis wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Protein adsorption wikipedia , lookup
Transcriptional regulation wikipedia , lookup
Polyadenylation wikipedia , lookup
Protein structure prediction wikipedia , lookup
Biochemistry wikipedia , lookup
Point mutation wikipedia , lookup
Metalloprotein wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Non-coding RNA wikipedia , lookup
Bottromycin wikipedia , lookup
Gene expression wikipedia , lookup
Expanded genetic code wikipedia , lookup
Messenger RNA wikipedia , lookup
Transfer RNA wikipedia , lookup
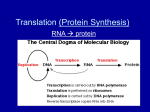
PROTEIN SYNTHESIS PROTEIN SYNTHESIS DNA: NUCLEIC ACID, DOUBLE STRAND, PO4, DEOXYRIBOSE SUGAR. RNA: NUCLEIC ACID, SINGLE STRAND, PO4, RIBOSE SUGAR. BASE PAIRS (N) T=THYMINE A=ADENINE C= CYTOSINE G=GUANINE BASE PAIRS (N) U = URACIL A=ADENINE C=CYTOSINE G=GUANINE URACIL (U) base with a single-ring structure phosphate group sugar (ribose) POINTS ABOUT TRANSCRIPTION NEED RNA POLYMERASE CODES FOR 20 AMINO ACIDS CODON:SERIES OF TRIPLET BASE PAIRS. 64 CODONS, 60 FOR AA, OTHERS FOR STARTS/STOPS. INTRONS=NON-CODING EXONS= CODING FOR RNA PROTEIN TRANSCRIPTION NUCLEUS RNA POLYMERASE CODES TO DNA DNA TRANSCRIBES TO m-RNA INTRONS SNIPPED OUT EXONS KEPT IN CODE exon unit of transcription in a DNA strand intron exon intron exon 3’ 5’ transcription into pre-mRNA poly-A tail cap 5’ 3’ (snipped out) (snipped out) 5’ 3’ mature mRNA transcript sugar-phosphate backbone of one strand of nucleotides in a DNA double helix sugar-phosphate backbone of the other strand of nucleotides part of the sequence of base pairs in DNA transcribed DNA winds up again DNA to be transcribed unwinds Newly forming RNA transcript The DNA template at the assembly site growing RNA transcript 5’ 3’ 5’ RNA polymerase direction of transcription 3’ 5’ 3’ PROTEIN TRANSLATION m-RNA GOES THRU RIBOSOME. RIBOSOME IS rRNA,CODE THREADS THRU RIBOSOME. AREA OF RIBOSOME BOUND TO tRNA 20 TYPES OF AA ANTICODON ON ONE END OF tRNA. AA ON OTHER END OF t-RNA AA ATTACH TO EACH OTHER IN PEPTIDE BOND FORM PROTEINS Binding site for mRNA P (first binding site for tRNA) A (second binding site for tRNA) TRANSCRIPTION Pre mRNA Transcript Processing Unwinding of gene regions of a DNA molecule mRNA rRNA tRNA protein subunits Mature mRNA transcripts TRANSLATION Synthesis of a polypetide chain at binding sites for mRNA and tRNA on the surface of an intact ribosome ribosomal subunits mature tRNA Convergence of RNAs Cytoplasmic pools of amino acids, tRNAs, and ribosomal subunits FINAL PROTEIN Destined for use in cell or for transport VALINE PROLINE THREONINE LEUCINE HISTIDINE GLUTAMATE GLUTAMATE VALINE PROLINE THREONINE VALINE LEUCINE HISTIDINE GLUTAMATE mRNA transcribed from the DNA PART OF PARENTAL DNA TEMPLATE ARGININE GLYCINE TYROSINE TRYPTOPHAN ASPARAGINE ARGININE GLYCINE LEUCINE LEUCINE GLUTAMATE resulting amino acid sequence altered message in mRNA A BASE INSERTION (RED) IN DNA the altered amino acid sequence Overview: the roles of transcription and translation in the flow of genetic information The triplet code TRANSCRIPTION AND TRANSLATION C DNA. m-RNA. t-RNA. AMINO ACID ATC-GCG-TAT UAG-CGC-AUA AUC-GCG-UAU ISO-ALA-TYR PEPTIDE BONDS/POLYPEPTIDES/PROTEINS Translation Nuclear membrane DNA Transcription Eukaryotic Cell Pre-mRNA RNA Processing mRNA Ribosome Translation Protein Translation Synthesis of proteins in the cytoplasm Involves the following: 1. mRNA (codons) 2. tRNA (anticodons) 3. rRNA 4. ribosomes 5. amino acids Types of RNA Three types of RNA: A. messenger RNA (mRNA) B. transfer RNA (tRNA) C. ribosome RNA (rRNA) Remember: all produced in the nucleus! A. Messenger RNA (mRNA) Carries the information for a specific protein. Made up of 500 to 1000 nucleotides long. Made up of codons (sequence of three bases: AUG - methionine). Each codon, is specific for an amino acid. A. Messenger RNA (mRNA) start codon mRNA A U G G G C U C C A U C G G C G C A U A A codon 1 protein methionine codon 2 codon 3 glycine serine codon 4 isoleucine codon 5 codon 6 glycine alanine codon 7 stop codon Primary structure of a protein aa1 aa2 aa3 peptide bonds aa4 aa5 aa6 B. Transfer RNA (tRNA) Made up of 75 to 80 nucleotides long. Picks up the appropriate amino acid floating in the cytoplasm (amino acid activating enzyme) Transports amino acids to the mRNA. Have anticodons that are complementary to mRNA codons. Recognizes the appropriate codons on the mRNA and bonds to them with H-bonds. anticodon codon in mRNA anticodon amino acid attachment site tRNA MOLECULE amino acid attachment site amino acid OH The structure of transfer RNA (tRNA) B. Transfer RNA (tRNA) amino acid attachment site methionine U A C anticodon amino acid C. Ribosomal RNA (rRNA) Made up of rRNA is 100 to 3000 nucleotides long. Important structural component of a ribosome. Associates with proteins to form ribosomes. Ribosomes Large and small subunits. Composed of rRNA (40%) and proteins (60%). Both units come together and help bind the mRNA and tRNA. Two sites for tRNA a. P site (first and last tRNA will attach) b. A site Ribosomes Origin Cytosol (eukaryotic ribosome) Chloroplasts (prokaryotic ribosome) Complete Ribosomal ribosome subunit 80 S 40 S 60 S rRNA components 18 S 5S 5.8 S 25 S Proteins 70 S 30 S 50 S 16 S 4.5 S 5 S 23 S C. 24 C. 35 30 S 50 S 18 S 5S 26 S C. 33 C. 35 Mitochondrion 78 S (prokaryotic ribosome) C.30 C.50 Ribosomes Large subunit P Site A Site mRNA A U G Small subunit C U A C U U C G Translation Three parts: 1. initiation: start codon (AUG) 2. elongation: 3. termination: stop codon (UAG) Let’s make a PROTEIN!!!!. Translation Large subunit P Site A Site mRNA A U G Small subunit C U A C U U C G Translation • Initiation The inactive 40S and 60S subunits will bind to each other with high affinity to form inactive complex unless kept apart This is achieved by eIF3, which bind to the 40S subunit mRNA forms an initiation complex with a ribosome A number of initiation factors participate in the process. 33 Translation Cap sequence present at the 5’ end of the mRNA is recognized by eIF4 Subsequently eIF3 is bound and cause the binding of small 40S subunit in the complexes The 18S RNA present in the 40 S subunit is involved in binding the cap sequence eIF2 binds GTP and initiation tRNA, which recognize the the start codon AUG This complex is also bound to 40S subunit 34 Translation Driven by hydrolysis of ATP, 40S complex migrate down stream until it finds AUG start codon The large 60S subunit is then bound to the 40S subunit It is accompanied by the dissociation of several initiation factor and GDP The formation of the initiation complex is now completed Ribosome complex is able to translate 35 Translation Extrachromosomal mRNAs have no cap site Plastid mRNA has a special ribosome binding site for the initial binding to the small subunit of the ribosome (shine-Dalgarno sequence) This sequence is also found in bacterial mRNA, but it is not known in the mitochondria In the prokaryotic, the initiation tRNA is loaded with N-formylmethionine After peptide formation, the formyl residue is cleaved from the methionine 36 Initiation aa1 aa2 2-tRNA 1-tRNA anticodon hydrogen bonds U A C A U G codon G A U C U A C U U C G A mRNA Translation • Elongation A ribosome contains two sites where the tRNAs can bind to the mRNA. P (peptidyl) site allows the binding of the initiation tRNA to the AUG start codon. The A (aminoacyl) site covers the second codon of the gene and the first is unoccupied On the other side of the P site is the exit (E) site where empty tRNA is released 38 Translation • Elongation The elongation begins after the corresponding aminoacyl-tRNA occupies the A site by forming base pairs with the second codon Two elongation factors (eEF) play an important role eEF1 binds GTP and guides the corresponding aminoacyl-tRNA to the A site, during which GTP is hydrolized to GDP and P. The cleavage of the energy-rich anhydride bond in GTP enables the aminoacyl-tRNA to bind to codon at the A site 39 Translation • Elongation Afterwards the GDP still bound to eEF1, is exchange for GTP as mediated by the eEF1 The eEF1 -GTP is now ready for the next cycle Subsequently a peptide linkage is form between the carboxyl group of methionine and the amino group of amino acid of the tRNA bound to A site Peptidyl transferase catalyzing the reaction. It facilitates the N-nucleophilic attack on the carboxyl group, whereby the peptide bond is formed with the released of water 40 Translation • Elongation Accompanied by the hydrolysis of one molecule GTP to form GDP and P, the eEF2 facilitates the translocation of the ribosome along the mRNA to three bases downstream Free tRNA arrives at site E is released, and tRNA loaded with the peptide now occupies the P Site The third aminoacyl-tRNA binds to the vacant A site and a further elongation cycle can begin 41 Elongation peptide bond aa3 aa1 aa2 3-tRNA 1-tRNA anticodon hydrogen bonds U A C A U G codon 2-tRNA G A A G A U C U A C U U C G A mRNA aa1 peptide bond aa3 aa2 1-tRNA 3-tRNA U A C (leaves) 2-tRNA A U G G A A G A U C U A C U U C G A mRNA Ribosomes move over one codon aa1 peptide bonds aa4 aa2 aa3 4-tRNA 2-tRNA A U G 3-tRNA G C U G A U G A A C U A C U U C G A A C U mRNA aa1 peptide bonds aa4 aa2 aa3 2-tRNA 4-tRNA G A U (leaves) 3-tRNA A U G G C U G A A C U A C U U C G A A C U mRNA Ribosomes move over one codon aa1 peptide bonds aa5 aa2 aa3 aa4 5-tRNA U G A 3-tRNA 4-tRNA G A A G C U G C U A C U U C G A A C U mRNA peptide bonds aa1 aa5 aa2 aa3 aa4 5-tRNA U G A 3-tRNA G A A 4-tRNA G C U G C U A C U U C G A A C U mRNA Ribosomes move over one codon aa4 aa5 Termination aa199 aa3 primary structure aa2 of a protein aa200 aa1 200-tRNA A C U mRNA terminator or stop codon C A U G U U U A G Translation • Release When A site finally binds to a stop codon (UGA, UAG, UAA) Stop codons bind eRF accompanied by hydrolysis GTP to form GDP and P Binding of eRF to the stop codon alters the specificity the peptidyl transferase Water instead amino acid is now the acceptor for the peptide chain Protein released from the tRNA Translation • The difference • Eukaryotic and prokaryotic translation can react differently to certain antibiotics Puromycin an analog tRNA and a general inhibitor of protein synthesis Cycloheximide only inhibits protein synthesis by eukaryotic ribosomes Chloramphenicol, Tetracycline, Streptomycin inhibit protein synthesis by prokaryotic ribosome End Product The end products of protein synthesis is a primary structure of a protein. A sequence of amino acid bonded together by peptide bonds. aa2 aa1 aa3 aa4 aa5 aa199 aa200 Polyribosome • Groups of ribosomes reading same mRNA simultaneously producing many proteins (polypeptides). incoming large subunit 1 incoming small subunit 2 3 4 polypeptide 5 6 7 mRNA TYPES OF PROTEINS ENZYMES/HELICASE CARRIER/HEMOGLOBIN IMMUNOGLOBULIN/ANTIBODIES HORMONES/STEROIDS STRUCTURAL/MUSCLE IONIC/K+,Na+ all regulate things put together ”critter” Protein Sorting Vast majority of protein within the cell are synthesized within the cytoplasm, but the final sub-cellular location can be in one of a whole array of membrane-bound compartment Protein is subjected to be sorted for special targeted organelles Protein Sorting Vast majority of protein within the cell are synthesized within the cytoplasm, but the final sub-cellular location can be in one of a whole array of membrane-bound compartment Protein is subjected to be sorted for special targeted organelles: Plastids Mitochondria Peroxisomes Vacuoles Mitochondria More than 95% of mitochondrial proteins in plant are encoded in the nucleus and translated in the cytosol Proteins are generally equipped with targeting signals ( a signal sequence of 12-70 amino acids at the amino terminal) Protein import occurs at translocation site In most cases, protein destined for the mitochondrial inner membrane after transport through outer membrane are guided directly to the location by internal targeting sequence Protein destined for the inner mitochondrial membrane contain prosequence that guides first into the mitochondrial matrix. After removal of the pro-sequence by processing peptidase, the proteins are directed by second targeting signal sequence into the inner membrane Plastids ATP is consumed for the phosphorilation of a protein, probably the receptor OEP86 The protein transport is regulated by the binding of the GTP to OEP86 and OEP34 After the protein is delivered, the pre-sequence is removed by a processing peptidase The protein destined to thylakoid membrane are first delivered into stroma and then directed by internal targeting signal into thylakoid membrane Peroxisomes Small membrane-bound cytoplasmic organelle containing oxidizing enzymes They can be found in leaf cells where they contain some of the enzymes of glycolytic pathway All protein have to be delivered from the cytosol The transport is accompanied by ATP hydrolysis Targeting sequence SKL (serine-lysine-leucine) has been observed in C terminus, but this sequence is not removed after uptake Vacuole Proteins are transferred during their synthesis to the lumen of ER This is aided by a signal sequence at the terminus of the synthesized protein, which binds with a signal recognition particle to a pore protein present in the ER membrane and thus directs the protein to the ER lumen In such cases, ribosome is attached to the ER membrane during protein synthesis and the synthesized protein appears immediately in the ER lumen. It is called co-translational protein transport This protein is then transferred from the ER by vesicles transfer across the golgi apparatus to the vacuole or are exported by secretory vesicles from the cell Coupled transcription and translation in bacteria original base triplet in a DNA strand a base substitution within the triplet (red) As DNA is replicated, proofreading enzymes detect the mistake and make a substitution for it: POSSIBLE OUTCOMES: OR One DNA molecule carries the original, unmutated sequence VALINE PROLINE The other DNA molecule carries a gene mutation THREONINE VALINE LEUCINE HISTIDINE GLUTAMATE A summary of transcription and translation in a eukaryotic cell