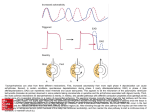
* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Practical Aspects of Estimating Energy Components in
Gene nomenclature wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Epitranscriptome wikipedia , lookup
Promoter (genetics) wikipedia , lookup
RNA interference wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Endogenous retrovirus wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Clinical neurochemistry wikipedia , lookup
Expression vector wikipedia , lookup
Biosynthesis wikipedia , lookup
Eukaryotic transcription wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Ultrasensitivity wikipedia , lookup
RNA polymerase II holoenzyme wikipedia , lookup
Signal transduction wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Secreted frizzled-related protein 1 wikipedia , lookup
Gene expression wikipedia , lookup
Transcriptional regulation wikipedia , lookup
Mitogen-activated protein kinase wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Amino acid synthesis wikipedia , lookup
Gene regulatory network wikipedia , lookup
Supplemental Text 1: SNP report of the rs2403254 locus. The report contains knowledge of all genes within a 500 kbp distance of rs2403254, as retrieved from NCBI-Gene, ConsensusPathDB, UniProtKB, OMIM, Gene Ontology, TCDB, ExPASy and KEGG database. -------------------------------------------------------------------------------------SNP rs2403254 Chr 11 18325146 -------------------------------------------------------------------------------------Distance to locus: 0 Gene: 11234 (HPS5) Chr 11 18300217-18343721 (-) 21833017 Seven mutations (six previously unreported) were described in the HPS1, HPS4, and HPS5 genes among Hermansky-Pudlak Syndrome patients of Mexican, Uruguayan, Honduran, Cuban, Venezuelan, and Salvadoran ancestries. 17301833 Tyrosinase and TYRP1 are mistrafficked, however, and fail to be efficiently delivered to melanosomes of HPS-5 melanocytes 15296495 LAMP-3 distribution was restricted to the perinuclear region in HPS-5 fibroblasts, instead of extending to the periphery 15030569 Component of BLOC-2. Results suggest a common biological basis underlying the pathogenesis of HPS-3, -5 and -6 disease. Protein: HPS5 (Q9UPZ3) Hermansky-Pudlak syndrome 5 protein; GO: 0006996 organelle organization GO: 0007596 blood coagulation GO: 0031084 BLOC-2 complex GO: 0043473 pigmentation OMIM: 607521 HPS5 GENE; HPS5 OMIM: 614074 HERMANSKY-PUDLAK SYNDROME 5; HPS5 Distance to locus: -18670 LD R2 of rs2403254 with intragenic SNP rs3802967: 1 Gene: 2965 (GTF2H1) Chr 11 18343816-18388590 (+) 21543505 These data suggest that the RVFV NSs protein is able to interact with the TFIIH subunit p62 inside infected cells and promotes its degradation, which can occur directly in the nucleus. 20974803 cdk1 phosphorylates p62 in vitro and in vivo at T269 and S272, which is necessary for the maintenance of appropriate cyclin B1 levels and the levels of cdk1 activity necessary to allow cells to properly enter and exit mitosis. 18692935 GTF2H1 polymorphisms/haplotypes may contribute to genetic susceptibility to lung cancer. 18270339 Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 15909982 The pleckstrin homology domain from the 62 kDa subunit Tfb1 (residues 1-108) of TFIIH is sufficient for binding to the activation domain of herpes simplex virus protein VP16. 15625236 p62 subunit of TFIIH interacts with TRbeta in a ligand-dependent manner. 14500720 transcription factor b3, previously identified as a component of the transcription factor IIH core complex, is shown instead to be transcription factor b4 Pathway: Nucleotide excision repair - Homo sapiens (human) (database: KEGG) Pathway: Basal transcription factors - Homo sapiens (human) (database: KEGG) Pathway: Viral carcinogenesis - Homo sapiens (human) (database: KEGG) Pathway: Eukaryotic Transcription Initiation (database: Wikipathways) Pathway: HIV Life Cycle (database: Reactome) Pathway: HIV Infection (database: Reactome) Pathway: RNA Polymerase I, RNA Polymerase III, and Mitochondrial Transcription (database: Reactome) Pathway: Transcription (database: Reactome) Pathway: Disease (database: Reactome) Pathway: Formation of incision complex in GG-NER (database: Reactome) Pathway: Dual incision reaction in GG-NER (database: Reactome) Pathway: Global Genomic NER (GG-NER) (database: Reactome) Pathway: Formation of transcription-coupled NER (TC-NER) repair complex (database: Reactome) Pathway: Dual incision reaction in TC-NER (database: Reactome) Pathway: Transcription-coupled NER (TC-NER) (database: Reactome) Pathway: Nucleotide Excision Repair (database: Reactome) Pathway: Formation of the HIV-1 Early Elongation Complex (database: PID) Pathway: mRNA Capping (database: Reactome) Pathway: Late Phase of HIV Life Cycle (database: Reactome) Pathway: AndrogenReceptor (database: NetPath) Pathway: RNA Polymerase I Transcription Initiation (database: Reactome) Pathway: DNA Repair (database: Reactome) Pathway: RNA Polymerase II Promoter Escape (database: Reactome) Pathway: RNA Polymerase I Chain Elongation (database: Reactome) Pathway: RNA Polymerase II Transcription Pre-Initiation And Promoter Opening (database: Reactome) Pathway: Formation of HIV-1 elongation complex in the absence of HIV-1 Tat (database: PID) Pathway: RNA Polymerase II Pre-transcription Events (database: PID) Pathway: RNA Pol II CTD phosphorylation and interaction with CE (database: Reactome) Pathway: Formation of the HIV-1 Early Elongation Complex (database: Reactome) Pathway: HIV Transcription Initiation (database: Reactome) Pathway: RNA Polymerase II HIV-1 Promoter Escape (database: PID) Pathway: RNA Polymerase II HIV Promoter Escape (database: Reactome) Pathway: Formation of HIV-1 elongation complex containing HIV-1 Tat (database: Reactome) Pathway: Tat-mediated elongation of the HIV-1 transcript (database: Reactome) Pathway: Formation of incision complex in GG-NER (database: PID) Pathway: HIV Transcription Elongation (database: Reactome) Pathway: RNA Polymerase II Transcription Elongation (database: PID) Pathway: Formation of HIV elongation complex in the absence of HIV Tat (database: Reactome) Pathway: Transcription of the HIV genome (database: Reactome) Pathway: mRNA Processing (database: Reactome) Pathway: RNA Polymerase I Promoter Clearance (database: PID) Pathway: RNA Polymerase I Promoter Clearance (database: Reactome) Pathway: Dual incision reaction in GG-NER (database: PID) Pathway: RNA Pol II CTD phosphorylation and interaction with CE (database: PID) Pathway: RNA Polymerase I Promoter Escape (database: PID) Pathway: Formation and Maturation of mRNA Transcript (database: PID) Pathway: Formation of HIV-1 elongation complex containing HIV-1 Tat (database: PID) Pathway: Formation of the Early Elongation Complex (database: PID) Pathway: RNA Polymerase II Pre-transcription Events (database: Reactome) Pathway: RNA Polymerase II Promoter Escape (database: PID) Pathway: Global Genomic NER (GG-NER) (database: PID) Pathway: Dual incision reaction in TC-NER (database: PID) Pathway: RNA Polymerase II Transcription Initiation (database: Reactome) Pathway: RNA Polymerase II Transcription Initiation And Promoter Clearance (database: Reactome) Pathway: HIV-1 Transcription Initiation (database: PID) Pathway: Transcription (database: PID) Pathway: HIV-1 Transcription Elongation (database: PID) Pathway: RNA Pol II CTD phosphorylation and interaction with CE (database: Reactome) Pathway: RNA Polymerase I Transcription (database: PID) Pathway: RNA Polymerase I Transcription Termination (database: PID) Pathway: Transcription-coupled NER (TC-NER) (database: PID) Pathway: RNA Polymerase I Promoter Escape (database: Reactome) Pathway: Formation of the Early Elongation Complex (database: Reactome) Pathway: RNA Polymerase II Transcription (database: PID) Pathway: Formation of RNA Pol II elongation complex (database: Reactome) Pathway: RNA Polymerase II Transcription Elongation (database: Reactome) Pathway: RNA Polymerase II Transcription (database: Reactome) Pathway: Gene Expression (database: Reactome) Pathway: mRNA Capping (database: PID) Pathway: Formation of transcription-coupled NER (TC-NER) repair complex (database: PID) Pathway: RNA Polymerase II Transcription Initiation (database: PID) Pathway: Tat-mediated elongation of the HIV-1 transcript (database: PID) Pathway: RNA Polymerase I Chain Elongation (database: PID) Pathway: RNA Polymerase I Transcription Initiation (database: PID) Pathway: RNA Polymerase II Transcription Initiation And Promoter Clearance (database: PID) Pathway: Formation of RNA Pol II elongation complex (database: PID) Pathway: RNA Polymerase II Transcription Pre-Initiation And Promoter Opening (database: PID) Pathway: RNA Pol II CTD phosphorylation and interaction with CE (database: PID) Pathway: RNA Polymerase I Transcription Termination (database: Reactome) Pathway: RNA Polymerase I Transcription (database: Reactome) Pathway: Transcription of the HIV genome (database: PID) Pathway: mRNA Processing (database: PID) Protein: TF2H1 (P32780) General transcription factor IIH subunit 1; GO: 0000079 regulation of cyclin-dependent protein serine/threonine kinase activity GO: 0000439 core TFIIH complex GO: 0000718 nucleotide-excision repair, DNA damage removal GO: 0005515 protein binding GO: 0005654 nucleoplasm GO: 0005675 holo TFIIH complex GO: 0006200 ATP catabolic process GO: 0006281 DNA repair GO: 0006283 transcription-coupled nucleotide-excision repair GO: 0006289 nucleotide-excision repair GO: 0006360 transcription from RNA polymerase I promoter GO: 0006361 transcription initiation from RNA polymerase I promoter GO: 0006362 transcription elongation from RNA polymerase I promoter GO: 0006363 termination of RNA polymerase I transcription GO: 0006366 transcription from RNA polymerase II promoter GO: 0006367 transcription initiation from RNA polymerase II promoter GO: 0006368 transcription elongation from RNA polymerase II promoter GO: 0006370 7-methylguanosine mRNA capping GO: 0006468 protein phosphorylation GO: 0010467 gene expression GO: 0016032 viral process GO: 0045944 positive regulation of transcription from RNA polymerase II promoter GO: 0050434 positive regulation of viral transcription KO: K03141 transcription initiation factor TFIIH subunit 1 OMIM: 189972 GENERAL TRANSCRIPTION FACTOR IIH, POLYPEPTIDE 1; GTF2H1 Distance to locus: 33622 LD R2 of rs2403254 with intragenic SNP rs12218: 0.62922 Gene: 6288 (SAA1) Chr 11 18287808-18291524 (+) 23623642 SAA up-regulates Lp-PLA2 production significantly via a FPRL1/MAPKs./PPAR-gamma signaling pathway. 23566403 SAA levels can be associated with clinical phenotypes in multiple sclerosis and neuromyelitis optica. 23454129 SAA stimulates foam cell formation via LOX1 induction, and thus likely contributes to atherogenesis. 23437051 SAA1 gene polymorphisms, consisting of -13T/C single nucleotide polymorphism (SNP) in the 5'-flanking region and SNPs within exon 3 are associated with susceptibility to Familial Mediterranean Fever in the Japanese population. 23391827 a novel STAT3 non-consensus TFBS at nt -75/-66 downstream of the NF-kappaB RE in the SAA1 promoter region that is required for NF-kappaB p65 and STAT3 to activate SAA1 transcription 23357645 variant p.Gly90Asp SAA1 protein eliciting significantly reduced inflammatory responses in macrophages through a reduction in the secretion of inflammatory cytokines. 23300020 High serum SAA level is associated with obesity and estrogen receptor-negative breast tumors. 23255027 Letter: serum amyloid A levels are elevated in venous thromboembolism. 23230824 in acute phase of epilepsy, the serum SAA increased, but after 3 months of treatment, the SAA peak was disappeared. 23223242 Pathogenic serum amyloid A 1.1 shows a long oligomer-rich fibrillation lag phase contrary to the highly amyloidogenic non-pathogenic SAA2.2 23171281 Renal AA-amyloidosis is the predominant cause of renal failure in intravenous drug users. Patients with AA-amyloidosis are more likely to be HIV infected and tend to have a higher rate of repeated systemic infections. 23165195 Lipid-poor SAA, but not high density lipoprotein-associated SAA, stimulates G-CSF production. 22901189 High serum amyloid A expression is associated with benign neoplasm of pancreas. 22827403 Data suggest that serum amyloid protein A (SAA1) level in gestational diabetes is predictive of subclinical atherosclerosis in these women; SAA1 levels correlate with risk factors and may serve as biomarker of future atherosclerotic heart disease. 22771314 SAA levels measured by ELISA in 97 NSCLC patients treated with gefitinib correlated with the clinical outcome of the patients 22768267 The rs12218 single nucleotide polymorphism in the SAA1 gene was associated with SUA levels in Chinese subjects, indicating that carriers of the T allele of rs12218 have a high risk of hyperuricemia. 22766792 Serum amyloid A-positive hepatocellular neoplasms may be a new type of inflammatory hepatocellular tumors in alcoholic patients. 22552174 In patients with persistent Atrial Fibrillation and preserved LVEF, serum amyloid a and CRP levels are independent predictors of AF subacute recurrence rate. 22447522 Serum amyloid A (SAA) induces pentraxin 3 (PTX3) production in rheumatoid synoviocytes. 22371903 Amyloid A (AA) proteins in biopsy samples taken from the stomachs of patients with AA amyloidosis 22367597 findings suggested that SAA was significantly associated with disease duration and gastrointestinal manifestations manifestations in Henoch-Schonlein purpura patients 22341353 Purified ten targeted proteins from sera of patients with various types of advanced stage(stage III-IV)cancers. These proteins were identified as isoforms of the human serum amyloid protein A (SAA) family with or without truncations at their N-terminals. 22300576 Large-scale isotype-specific quantification of Serum amyloid A 1/2 by multiple reaction monitoring in crude sera. 22076945 acute phase SAA may be involved in joint destruction though MMP induction and collagen cleavage in vivo. 22011672 Reactive amyloid A (AA) amyloidosis, one of the most severe complications of RA, is a serious, potentially life-threatening disorder caused by deposition of AA amyloid fibrils in multiple organs. 21953030 report abundance levels of SAA in serum samples from patients with advanced breast cancer, colorectal cancer (CRC) and lung cancer compared to healthy controls (age and gender matched) using commercially available enzyme-linked immunosorbent assay kits 21876610 Serum amyloid alpha is increased in complicated parapneumonic effusion, and it might be useful as a biomarker for complicated and uncomplicated parapneumonic effusion. 21624307 The genetic polymorphism rs12218 of SAA1 might be a promoting factor for carotid intima media thickness in Han Chinese people. 21569756 the N-terminal region plays a crucial role as a rigid core and the central region facilitates the elongation of fibrils in heparin-induced amyloidogenesis of SAA molecule 21465531 SAA up-regulates PTX3 production via FPRL1 significantly, and thus, contributes to the inflammatory pathogenesis of atherosclerosis. 21449704 CC genotype of rs12218 in the SAA1 gene was associated with decreased ABI in Chinese Han subjects, which indicated that the carriers of CC genotype of rs12218 have high risk of peripheral arterial disease. 21325601 A novel anti-inflammatory role of SAA in the induction of a micro-environment that supports T(reg) expansion at sites of infection or tissue injury, likely to curb (auto)-inflammatory responses. 21309051 Gestational diabetes is associated with loss of natural variation of C-reactive protein and serum amyloid A, suggesting altered modulation of inflammation. 21141971 Data suggest that a higher concentration of SAA can serve as an indicator of lung adenocarcinoma and represents a therapeutic target for the inhibition of lung cancer metastasis. 21124955 These findings demonstrate a key role of the 11p15.5-p13 region in the regulation of baseline A-SAA levels. 21103356 Data show that both rs12218 of the SAA1 gene and rs2468844 of SAA2 gene are associated with carotid IMT in healthy Han Chinese subjects. 21067563 Intestinal epithelial SAA displays bactericidal properties in vitro and could play a protective role in experimental mouse colitis. 20979827 In Chinese Han people, the genetic polymorphism of SAA1 may reflect the serum concentration of HDL-C. 20964562 The blood levels of serum amyloid A protein and transthyretin were found to be significantly increased in lung cancer patients. 20877498 Serum amyloid A protein was significantly increased in vivax malaria patient's plasma. 20869715 acute-phase Serum Amyloid A has a role in response to metformin treatment in insulin resistant women with the polycystic ovary syndrome 20853849 This study data suggested that SAA activated PPARgamma through extracellular signal-regulated kinase 1/2 (ERK1/2)-dependent COX-2 expression. 20713982 the expression of SAA in human benign and malignant ovarian epithelial tumors 20502455 A differential protein with m/z 11.6 kDa was detected and identified as an isoform of human serum amyloid A (SAA). It was significantly increased by 1822% in lung cancer patients when compared with the healthy controls. 20463814 Induction of acute phase protein serum amyloid A (A-SAA) occurred as early as 5-7 days prior to the first detection of plasma viral HIV-1 RNA, considerably prior to any elevation in systemic cytokine levels 20435930 A-SAA promotes cell migrational mechanisms and angiogenesis critical to rehumatoid arthritis pathogenesis. 20184536 The infectious group was subdivided into bacterial and viral infections, significantly higher leukocyte, serum amyloid A values were observed. 20177146 The results indicate that SAA stimulates FPR2-mediated activation of p38 MAPK and JNK, which are independent of a pertussis toxin-sensitive G-protein and are essential for SAA-induced CCL2 production. 20061576 Data indicate that SAA1 contributes to the reduced reverse cholesterol transport observed during acute phase responses 20022911 High SAA1 is associated with metastatic renal cell cancer. 19925664 Measuring the oxidation-reduction potential (ORP) of plasma in the critically injured, with injury severity and serum amyloid A (SAA) levels. Results suggest the presence of an oxidative environment in the plasma of critically injured as measured by ORP. 19922330 Results suggest that the relatively high affinity of SAA1.5 for high-density lipoprotein may cause the high serum concentration and may be related to the low susceptibility to amyloidosis. 19850938 During atherogenesis, SAA can amplify the involvement of smooth muscle cells in vascular inflammation and that this can lead to deposition of sPLA(2) and subsequent local changes in lipid homeostasis. 19833744 Results describe a significant inverse correlation between serum amyloid A and IgG levels and anti-SSA antibody titers as well as a trend towards an inverse correlation between SAA and ANA and RF titers, but not between CRP and these autoantibodies. 19758820 serum levels of SAA are strongly correlated with serum levels of inflammatory markers as well as measures of glycemic control 19755416 hepatic serum amyloid A expression is enhanced via protein kinase A-dependent mechanism 19729864 Associations of SAA1 and SAAP2 SNPs with carotid artery intima-media thickness, HDL and total cardiovascular disease events 19726303 SAA levels are elevated in obstructive sleep apnea syndrome patients in close correlation to the severity of OSAS, which may contribute to the vulnerability of the patients to cardiovascular diseases 19549924 a highly conserved motif required for SAA binding to macrophages can, under acidic pH conditions and in an heparan sulfate -dependent manner, also act as a molecular switch, directing SAA misfolding into AA amyloid. 19453393 High expression of SAA in carcinomas was detected mostly in tumor cells, but not in normal mucosa. AG490, inhibitor of STAT3 activation, reduced SAA1 expression in SAS cells. SAA was up-regulated in HNSCC through the Janus kinase-STAT3 pathway 19171342 haptoglobin and serum amyloid A may have roles in atherothrombotic ischemic stroke 19167353 SAA stimulates CCL2 production via FPRL1 and, thus, contributes to atherosclerosis. 19007930 A novel oxidized low-density lipoprotein marker, serum amyloid A-LDL, is associated with obesity and the metabolic syndrome. 18768891 SAA, an endogenous ligand of the chemokine formyl peptide receptor-like 1 (FPRL1), stimulates CCL2 production in human monocytes by activating extracellular-signal regulated kinase (ERK) and NF-kappaB. 18726069 A review of how SAA contributes to tumour development and the acceleration of tumour progression and metastasis is presented. 18695905 up-regulation of SAA protein expression mediates the inhibitory effect of Ganoderma lucidum polysaccharides on tumor cell adhesion to endothelial cells. 18571179 SAA is a prothrombotic and proinflammatory mediator in acute coronary syndrome which may contribute to atherogenesis and its complications 18566366 suggest a potential role for SAA in inflammatory diseases through activation of TLR2 18385816 fifth day of observation but were not good predictors of mortality in septic shock. 18322992 The SAA-RAGE-stimulated NF-kappaB signaling pathway has an important role in the pathogenesis of rheumatoid arthritis. 18299466 The association between SAA and leptin suggests an interaction between these two adipokines, which may have implications in inflammatory processes related to obesity and the metabolic syndrome 17968686 Analysis of SAA1 gene polymorphisms showed the rarity of the putative amyloidogenic -13T allele in Greek populations may be related to low prevalence of AA amyloidosis development in Greek RA patients. 17849429 we first show that SAA1 (coding for the major SAA isoform) but not SAA2 transcripts are expressed in human trabecular and cortical bone fractions and bone marrow. 17806085 SSA1 protein was identified in the blood of colon neoplasm patients. 17676666 SAA was quantitated using PVDF affinity probes and MALD-MS. 17552057 There is a relative deficiency of circulating cystatin C (CysC) in systemic inflammation in rheumatoid arthritis. Interaction between CysC and serum amyloid A protein explains this CysC deficiency and suggests CysC is regulating inflammatory responses. 17461519 SAA can be used as a valuable indicator of disease activity in ankylosing spondylitis. 17329325 SAA reduces hepatitis C virus (HCV) infectivity in a dose-dependent manner when added during HCV infection but not after virus entry. 17237436 SAA induced TF mRNA in PBMC within 30 min and optimal procoagulant activity within 4 h through activation of NF-kappaB via the ERK1/2 and p38 MAPK pathways. SAA-induced TF was partially inhibited by high-density lipoprotein. 17039260 the SAA1 alpha/alpha genotype is a risk factor for amyloidosis in BD 17015746 binding of SAA to FPRL1 may contribute to the destruction of bone and cartilage via the promotion of synoviocyte hyperplasia and angiogenesis 16864904 serum amyloid A and C-reactive protein are expressed differently in exudates and transudates 16737350 Increased expression of SAA1 by adipocytes in obesity may play a critical role in local and systemic inflammation and free fatty acid production and could be a direct link between obesity and its comorbidities. 16651021 Data show that serum amyloid A dissociates apolipoprotein E from high density lipoprotein in cerebrospinal fluid. 16236134 We provide evidence that STAT3 plays an essential role in cytokine-driven SAA expression, although the human SAA gene shows no typical STAT3 response element. 16155472 Within a subsample from the BELSTRESS study of 892 male subjects free of cardiovascular disease, dimensions of job stress from the job demand-control-support model were related to biomarkers of inflammation and infection. 16120612 Serum amyloid A has a role in promoting cholesterol efflux mediated by scavenger receptor B-I 15972323 Increased amyloid AA is associated with amyloidosis and familial Mediterranean fever 15910745 In this study, the SAA4 protein was examined in the high-density lipoprotein fraction of both healthy and diseased individuals. 15729583 SAA is expressed by subcutaneous white adipose tissue and its production at this site is regulated by nutritional status. 15561721 SR-BI plays a key role in SAA metabolism through its ability to interact with and internalize SAA and SAA influences HDL cholesterol metabolism through its inhibitory effects on SR-BI-mediated selective lipid uptake 15225640 Human hepatocytes stimulated by Lipopolysaccharide produced serum amyloid A protein. 15188355 The up-regulation of the A-SAA and FPRL1 genes in inflamed synovial tissue suggests an important role in the pathophysiology of inflammatory arthritis. 15170927 SAA1a/a genotype is one genetic factor that confers a significant risk for amyloidosis in the Turkish FMF population but neither SAA1 nor SAA2 genotypes had a significant effect on SAA level. 15161744 Positive relationship between Tanis mRNA and the acute-phase protein serum amyloid A suggests an interaction between innate immune system responses and Tanis expression in muscle and adipose tissue. 14871291 Saa1 is regulated by tumor necrosis factor-alpha, interleukin-6 and glucocorticoids in hepatic and epithelial cells 14738910 Results indicate a novel role for interleukin-18 in rheumatoid inflammation through the synovial serum amyloid A production. 12762136 Data show higher SAA1 homozygosity in familial Mediterranean fever (FMF)-amyloidosis patients than in FMF patients, but no significant difference between controls and FMF patients with and without amyloidosis for the TNF-alpha-308 G-A allele. 12762135 Data suggest that the frequency of the -13T serum amyloid A1 (SAA1) allele may explain the difference in prevalence of AA amyloidosis in Japanese and Caucasians. 12410800 the region containing the C/EBPalpha,beta consensus binding site between the bases -252 and -175 is important for the glucocorticoid-induced SAA1 gene expression in HASMCs but not in HepG2 cells 12077270 The endogenous SAA1 gene has a cytokine-driven transcriptional disadvantage compared to SAA2 that is superseded by a marginal transcriptional advantage when glucocorticoids are present. 12056504 relation to cholesterol metabolism and cardiovascular disease (REVIEW) 11830469 human platelets specifically adhere to SAA in an RGD- and alphaIIbbeta3-dependent manner, indicating a role for SAA in modulating platelet adhesion Pathway: Selenium Pathway (database: Wikipathways) Pathway: Vitamin B12 Metabolism (database: Wikipathways) Pathway: Folate Metabolism (database: Wikipathways) Pathway: TRIF-mediated TLR3/TLR4 signaling (database: Reactome) Pathway: MyD88-independent cascade (database: Reactome) Pathway: Activated TLR4 signalling (database: Reactome) Pathway: Toll Like Receptor 4 (TLR4) Cascade (database: Reactome) Pathway: Scavenging by Class B Receptors (database: Reactome) Pathway: Toll Like Receptor 9 (TLR9) Cascade (database: Reactome) Pathway: MyD88 cascade initiated on plasma membrane (database: Reactome) Pathway: Toll Like Receptor 10 (TLR10) Cascade (database: Reactome) Pathway: Toll Like Receptor 3 (TLR3) Cascade (database: Reactome) Pathway: Toll Like Receptor 5 (TLR5) Cascade (database: Reactome) Pathway: TRAF6 Mediated Induction of proinflammatory cytokines (database: Reactome) Pathway: Toll Like Receptor 7/8 (TLR7/8) Cascade (database: Reactome) Pathway: Formyl peptide receptors bind formyl peptides and many other ligands (database: Reactome) Pathway: Peptide ligand-binding receptors (database: Reactome) Pathway: Toll-Like Receptors Cascades (database: Reactome) Pathway: TRAF6 mediated NF-kB activation (database: Reactome) Pathway: GPCR ligand binding (database: Reactome) Pathway: DEx/H-box helicases activate type I IFN and inflammatory cytokines production (database: Reactome) Pathway: Cytosolic sensors of pathogen-associated DNA (database: Reactome) Pathway: Toll Like Receptor 2 (TLR2) Cascade (database: Reactome) Pathway: Innate Immune System (database: Reactome) Pathway: ZBP1(DAI) mediated induction of type I IFNs (database: Reactome) Pathway: MyD88:Mal cascade initiated on plasma membrane (database: Reactome) Pathway: Toll Like Receptor TLR1:TLR2 Cascade (database: Reactome) Pathway: Toll Like Receptor TLR6:TLR2 Cascade (database: Reactome) Pathway: Immune System (database: Reactome) Pathway: G alpha (q) signalling events (database: Reactome) Pathway: Signal Transduction (database: Reactome) Pathway: Gastrin-CREB signalling pathway via PKC and MAPK (database: Reactome) Pathway: GPCR downstream signaling (database: Reactome) Pathway: RIP-mediated NFkB activation via ZBP1 (database: Reactome) Pathway: TAK1 activates NFkB by phosphorylation and activation of IKKs complex (database: Reactome) Pathway: G alpha (i) signalling events (database: Reactome) Pathway: TRAF6 mediated induction of NFkB and MAP kinases upon TLR7/8 or 9 activation (database: Reactome) Pathway: MyD88 dependent cascade initiated on endosome (database: Reactome) Pathway: Class A/1 (Rhodopsin-like receptors) (database: Reactome) Pathway: Advanced glycosylation endproduct receptor signaling (database: Reactome) Pathway: RIG-I/MDA5 mediated induction of IFN-alpha/beta pathways (database: Reactome) Pathway: Endogenous TLR signaling (database: PID) Pathway: Binding and Uptake of Ligands by Scavenger Receptors (database: Reactome) Pathway: Signaling by GPCR (database: Reactome) Protein: SAA1 (P0DJI8) Serum amyloid A-1 protein; GO: 0001664 G-protein coupled receptor binding GO: 0005576 extracellular region GO: 0006953 acute-phase response GO: 0007204 positive regulation of cytosolic calcium ion concentration GO: 0030168 platelet activation GO: 0030593 neutrophil chemotaxis GO: 0034364 high-density lipoprotein particle GO: 0045087 innate immune response GO: 0045785 positive regulation of cell adhesion GO: 0048246 macrophage chemotaxis GO: 0048247 lymphocyte chemotaxis GO: 0050708 regulation of protein secretion GO: 0050715 positive regulation of cytokine secretion GO: 0050716 positive regulation of interleukin-1 secretion GO: 0050728 negative regulation of inflammatory response GO: 0070062 extracellular vesicular exosome GO: 0071682 endocytic vesicle lumen KO: K17310 serum amyloid A protein OMIM: 104750 SERUM AMYLOID A1; SAA1 Distance to locus: 40098 Gene: 144106 (ST13P5) LD R2 of rs2403254 with intragenic SNP rs10832913: 0.40783 Chr 11 18283432-18285048 (+) Distance to locus: -54925 LD R2 of rs2403254 with intragenic SNP rs1520886: 0.024498 Gene: 100528017 (SAA2-SAA4) Chr 11 18252902-18270221 (-) Distance to locus: -54925 LD R2 of rs2403254 with intragenic SNP rs1520886: 0.024498 Gene: 6289 (SAA2) Chr 11 18259780-18270221 (-) 23223242 Pathogenic serum amyloid A 1.1 shows a long oligomer-rich fibrillation lag phase contrary to the highly amyloidogenic non-pathogenic SAA2.2 22300576 successful quantification of SAA2 in crude serum by MRM, for the first time, shows that SAA2 can be a good biomarker for the detection of lung cancers. 21103356 Data show that both rs12218 of the SAA1 gene and rs2468844 of SAA2 gene are associated with carotid IMT in healthy Han Chinese subjects. 20379614 Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 18385816 CRP and SAA strongly correlated up to the fifth day of observation but were not good predictors of mortality in septic shock. 16737350 Increased expression of SAA2 by adipocytes in obesity may play a critical role in local and systemic inflammation and free fatty acid production and could be a direct link between obesity and its comorbidities. 15170927 SAA1a/a genotype is one genetic factor that confers a significant risk for amyloidosis in the Turkish FMF population but neither SAA1 nor SAA2 genotypes had a significant effect on SAA level. 14871291 saa2 is regulated by tumor necrosis factor-alpha, interleukin-6, and glucocorticoids in hepatic and epithelial cells. 12077270 The glucocorticoid response element of the SAA2 promoter is dysfunctional compared to that of SAA1, hence glucocorticoids are unable to enhance the cytokine-driven transcriptional activity of SAA2. Pathway: Selenium Pathway (database: Wikipathways) Pathway: Vitamin B12 Metabolism (database: Wikipathways) Pathway: Folate Metabolism (database: Wikipathways) Pathway: TRIF-mediated TLR3/TLR4 signaling (database: Reactome) Pathway: MyD88-independent cascade (database: Reactome) Pathway: Activated TLR4 signalling (database: Reactome) Pathway: Toll Like Receptor 4 (TLR4) Cascade (database: Reactome) Pathway: Scavenging by Class B Receptors (database: Reactome) Pathway: Toll Like Receptor 9 (TLR9) Cascade (database: Reactome) Pathway: MyD88 cascade initiated on plasma membrane (database: Reactome) Pathway: Toll Like Receptor 10 (TLR10) Cascade (database: Reactome) Pathway: Toll Like Receptor 3 (TLR3) Cascade (database: Reactome) Pathway: Toll Like Receptor 5 (TLR5) Cascade (database: Reactome) Pathway: TRAF6 Mediated Induction of proinflammatory cytokines (database: Reactome) Pathway: Toll Like Receptor 7/8 (TLR7/8) Cascade (database: Reactome) Pathway: Formyl peptide receptors bind formyl peptides and many other ligands (database: Reactome) Pathway: Peptide ligand-binding receptors (database: Reactome) Pathway: Toll-Like Receptors Cascades (database: Reactome) Pathway: TRAF6 mediated NF-kB activation (database: Reactome) Pathway: GPCR ligand binding (database: Reactome) Pathway: DEx/H-box helicases activate type I IFN and inflammatory cytokines production (database: Reactome) Pathway: Cytosolic sensors of pathogen-associated DNA (database: Reactome) Pathway: Toll Like Receptor 2 (TLR2) Cascade (database: Reactome) Pathway: Innate Immune System (database: Reactome) Pathway: ZBP1(DAI) mediated induction of type I IFNs (database: Reactome) Pathway: MyD88:Mal cascade initiated on plasma membrane (database: Reactome) Pathway: Toll Like Receptor TLR1:TLR2 Cascade (database: Reactome) Pathway: Toll Like Receptor TLR6:TLR2 Cascade (database: Reactome) Pathway: Immune System (database: Reactome) Pathway: G alpha (q) signalling events (database: Reactome) Pathway: Signal Transduction (database: Reactome) Pathway: Gastrin-CREB signalling pathway via PKC and MAPK (database: Reactome) Pathway: GPCR downstream signaling (database: Reactome) Pathway: RIP-mediated NFkB activation via ZBP1 (database: Reactome) Pathway: TAK1 activates NFkB by phosphorylation and activation of IKKs complex (database: Reactome) Pathway: G alpha (i) signalling events (database: Reactome) Pathway: TRAF6 mediated induction of NFkB and MAP kinases upon TLR7/8 or 9 activation (database: Reactome) Pathway: MyD88 dependent cascade initiated on endosome (database: Reactome) Pathway: Class A/1 (Rhodopsin-like receptors) (database: Reactome) Pathway: Advanced glycosylation endproduct receptor signaling (database: Reactome) Pathway: RIG-I/MDA5 mediated induction of IFN-alpha/beta pathways (database: Reactome) Pathway: Endogenous TLR signaling (database: PID) Pathway: Binding and Uptake of Ligands by Scavenger Receptors (database: Reactome) Pathway: Signaling by GPCR (database: Reactome) Protein: SAA2 (P0DJI9) Serum amyloid A-2 protein; GO: 0006953 acute-phase response GO: 0034364 high-density lipoprotein particle KO: K17310 serum amyloid A protein OMIM: 104751 SERUM AMYLOID A2; SAA2 Distance to locus: -66762 LD R2 of rs2403254 with intragenic SNP rs2460827: 0.010826 Gene: 6291 (SAA4) Chr 11 18252902-18258384 (-) 20536400 studies provide the first evidence for a novel type of AA amyloidosis in which the fibrils were formed from a mutated SAA4 protein 12410800 The sequences of 1478 and 1836 bp of the SAA1 and SAA4 5'-flanking regions were determined. Pathway: Selenium Pathway (database: Wikipathways) Pathway: Vitamin B12 Metabolism (database: Wikipathways) Pathway: Folate Metabolism (database: Wikipathways) Protein: SAA4 (P35542) Serum amyloid A-4 protein; GO: 0005576 extracellular region GO: 0006953 acute-phase response GO: 0034364 high-density lipoprotein particle KO: K17310 serum amyloid A protein OMIM: 104752 SERUM AMYLOID A4; SAA4 Distance to locus: -84188 Gene: 100423016 (MIR3159) Chr 11 18409334-18409407 (+) Distance to locus: 90021 LD R2 of rs2403254 with intragenic SNP rs2468790: 0.017585 Gene: 494141 (LOC494141) Chr 11 18230685-18235125 (+) Distance to locus: -90790 LD R2 of rs2403254 with intragenic SNP rs3758683: 0.11936 Gene: 3939 (LDHA) Chr 11 18415936-18429765 (+) 23516535 the role played by ERRalpha in the regulation of lactate dehydrogenases A and B 23404405 Lactate dehydrogenase A is overexpressed in pancreatic cancer. 23266049 Data indicate that serum lactic dehydrogenase (S-LDH) appears to be a significant independent prognostic index in patients with metastatic nasopharyngeal carcinoma (NPC). 23184277 Increased LDH5 expression is associated with lymph node metastasis in oral squamous cell carcinoma. 23166385 Elevated lactate dehydrogenase level is associated with recurrent or refractory aggressive lymphoma. 22961700 LDHA plays an important role in the progression of esophageal squamous cell carcinoma by modulating cell growth 22948140 Cells expressing either PDK1 or LDHA maintained a lower mitochondrial membrane potential and decreased reactive oxygen species production with or without exposure to toxins. 22923663 Lactic acid induces myofibroblast differentiation via pH-dependent activation of transforming growth factor beta. 22897481 Studies indicate the mechanisms by which lactate dehydrogenase A (LDHA) promotes tumor growth and metastasis. 22593701 Data suggest that serum lactate dehydrogenase (LDH) kinetics might reflect disease behaviour in extracranial metastatic and primary sites without need for comprehensive imaging studies and is a quite inexpensive diagnostic test. 22429998 We demonstrate that LDH-A reduction can suppress the tumorigenicity of intestinal-type gastric cancer (ITGC) cells by downregulating Oct4 both in vitro and in vivo. 22360420 A protein encoded by this locus was found to be differentially expressed in postmortem brains from patients with atypical frontotemporal lobar degeneration. 22127970 Case Report: describe an elderly patient with physical therapy-induced rhabdomyolysis complicated by acute kidney injury associated with reduced skeletal muscle LDH-A activity. 21969607 LDH-A is tyrosine phosphorylated and activated by FGFR1 in cancer cells. 21858537 Results show that S-100B, MIA and LDH levels were significantly higher in patients with advanced melanoma than in disease-free patients or healthy controls. 21632858 Serum LDH and tissue LDH5 levels are complementary features that help to characterize the activity of LDH in colorectal cancer and have a potent value in predicting response to chemotherapy. 21452021 LDH-A reduction resulted in an inhibited cancer cell proliferation, elevated intracellular oxidative stress, and induction of mitochondrial pathway apoptosis. 21249322 High LDH is associated with M1b prostate cancer. 20951115 Presented are QM/MM calculations that show differences in geometries of active sites of M(4) and H(4) isoforms of human LDH ligated with oxamate, pyruvate or L-lactate. 20828817 High lactate dehydrogenase is associated with acute adult T-cell leukemia/lymphoma. 20385008 LDH5 is overexpressed in non-small cell lung cancer and could serve as a marker for malignancy. LDH5 correlates positively with the prognostic marker transketolase like 1 protein. 19923867 LDH5 is highly expressed in squamous cell head and neck cancer and is linked with local relapse, survival and distant metastasis. 19847924 Observational study and genome-wide association study of gene-disease association. (HuGE Navigator) 19838163 Correlation of LDH-5 expression with clinicopathological factors and with the expression of Bcl-2, Bcl-XL, Mcl-1 and GRP78 was examined in pigmented lesions, including nevi and melanoma at different stages of progression 19668225 ErbB2 promotes glycolysis at least partially through the HSF1-mediated upregulation of LDH-A. 19276158 LDH-A knockdown in the background of FH knockdown results in significant reduction in tumor growth in a xenograft mouse model. 19021062 LDH5 is highly upregulated in B-cell non-Hodgkin lymphomas and is in direct relation to factor HIF1alpha and HIF2alpha expression. LDH5 expression is linked with activated VEGFR2/KDR expression in both lymphoid lesions. 18821170 The expression of LDH and its isoenzymes in pleural effusions reflects the host reaction in pleural space and, in non-small-cell lung cancer, may also feature the anaerobic phenotype of cancer cells. 18814027 Modulation of LDH expression involves alpha6beta4 integrin-FAK-p38MAPK pathway. 18534967 LDL-M is released into blood fo patients exposed to myocardial ischemia reperfusion. 18521687 The results of the current study show that LDH-5 expression may be a useful prognostic factor for patients with gastric carcinoma. 17483170 biophysical study of ligand binding and protein dynamics in lactate dehydrogenase 17178662 LDH1 was decreased in essential thrombocythemia. This isoenzymatic pattern could be expression of a metabolic adaptation. 17178662 LDH5 was reduced in idiopathic myelofibrosis. This isoenzymatic pattern could be expression of a metabolic adaptation [LDH5] 16766262 Reduction in LDH-A activity resulted in stimulation of mitochondrial respiration and decrease of mitochondrial membrane potential.The tumorigenicity of the LDH-A-deficient cells was severely diminished. 16132575 Lactate dehydrogenase 5 content in tumor cells is directly related to an up-regulated hypoxia inducible factor pathway and is linked with an aggressive phenotype in colorectal adenocarcinomas. 15240094 These data indicate that LDH-A is induced through a non-genomic pathway of estrogen action. 12712614 The activity of this enzyme was studied in tissues, erythrocytes, and blood plasma of patients with peptic ulcer both in its uncomplicated course and in the development of complications. 12629811 The study of this protein in a sportsman is significant for assessment of training efficiency. 12555229 an LDHA exon5 haplotype confers increased risk for paradoxically decreased minute volume respiratory response to CO2 challenge but not to panic disorder Pathway: Pyruvate metabolism - Homo sapiens (human) (database: KEGG) Pathway: Glycolysis / Gluconeogenesis - Homo sapiens (human) (database: KEGG) Pathway: Propanoate metabolism - Homo sapiens (human) (database: KEGG) Pathway: HIF-1 signaling pathway - Homo sapiens (human) (database: KEGG) Pathway: Cysteine and methionine metabolism - Homo sapiens (human) (database: KEGG) Pathway: Cori Cycle (database: Wikipathways) Pathway: Glycolysis and Gluconeogenesis (database: Wikipathways) Pathway: hypoxia-inducible factor in the cardivascular system (database: BioCarta) Pathway: TCR (database: NetPath) Pathway: Pyruvate metabolism (database: Reactome) Pathway: Validated targets of C-MYC transcriptional activation (database: PID) Pathway: Pyruvate metabolism and Citric Acid (TCA) cycle (database: Reactome) Pathway: The citric acid (TCA) cycle and respiratory electron transport (database: Reactome) Pathway: Methionine Cysteine metabolism (database: INOH) Pathway: Propanoate metabolism (database: INOH) Pathway: Glycolysis Gluconeogenesis (database: INOH) Pathway: EGFR1 (database: NetPath) Pathway: Pyruvate metabolism (database: INOH) Pathway: pyruvate fermentation to lactate (database: HumanCyc) Pathway: HIF-1-alpha transcription factor network (database: PID) Pathway: hypoxia-inducible factor in the cardivascular system (database: PID) Protein: LDHA (P00338) L-lactate dehydrogenase A chain; EC: 1.1.1.27 L-lactate dehydrogenase. (S)-lactate + NAD(+) = pyruvate + NADH. GO: 0004459 L-lactate dehydrogenase activity GO: 0005515 protein binding GO: 0005634 nucleus GO: 0005739 mitochondrion GO: 0005829 cytosol GO: 0005929 cilium GO: 0006090 pyruvate metabolic process GO: 0006096 glycolytic process GO: 0021762 substantia nigra development GO: 0031668 cellular response to extracellular stimulus GO: 0044237 cellular metabolic process GO: 0044281 small molecule metabolic process GO: 0070062 extracellular vesicular exosome KO: K00016 L-lactate dehydrogenase [EC:1.1.1.27] ReactionKEGG: R00703 (S)-Lactate + NAD+ <=> Pyruvate + NADH + H+ ReactionKEGG: R01000 2-Hydroxybutanoic acid + NAD+ <=> 2-Oxobutanoate + NADH + H+ ReactionKEGG: R03104 3-Mercaptolactate + NAD+ <=> Mercaptopyruvate + NADH + H+ OMIM: 150000 LACTATE DEHYDROGENASE A; LDHA OMIM: 612933 GLYCOGEN STORAGE DISEASE XI; GSD11 Distance to locus: 106254 LD R2 of rs2403254 with intragenic SNP rs2460832: 0.015713 Gene: 645319 (LOC645319) Chr 11 18218207-18218892 (+) Distance to locus: -108707 LD R2 of rs2403254 with intragenic SNP rs7938942: 0.089432 Gene: 3948 (LDHC) Chr 11 18433853-18472793 (+) 21858537 Results show that S-100B, MIA and LDH levels were significantly higher in patients with advanced melanoma than in disease-free patients or healthy controls. 20499337 Elevated serum LDH isoenzymes and AST indicate a disturbance (of uncertain clinical significance) within multiple extraosseous tissues when there is CLCN7 deficiency. 18930904 hLdhc expression in cancer cells was regulated by transcription factor Sp1 and CREB and promoter CGI methylation 17935709 LDH3 is a supporting diagnostic marker in cases of chronic tuberculosis. 17178662 LDH3 was increased in essential thrombocythemia. This isoenzymatic pattern could be expression of a metabolic adaptation. Pathway: Pyruvate metabolism - Homo sapiens (human) (database: KEGG) Pathway: Glycolysis / Gluconeogenesis - Homo sapiens (human) (database: KEGG) Pathway: Propanoate metabolism - Homo sapiens (human) (database: KEGG) Pathway: Cysteine and methionine metabolism - Homo sapiens (human) (database: KEGG) Pathway: Glycolysis and Gluconeogenesis (database: Wikipathways) Pathway: Methionine Cysteine metabolism (database: INOH) Pathway: Propanoate metabolism (database: INOH) Pathway: Glycolysis Gluconeogenesis (database: INOH) Pathway: Pyruvate metabolism (database: INOH) Pathway: pyruvate fermentation to lactate (database: HumanCyc) Protein: LDHC (P07864) L-lactate dehydrogenase C chain; EC: 1.1.1.27 L-lactate dehydrogenase. (S)-lactate + NAD(+) = pyruvate + NADH. GO: 0004459 L-lactate dehydrogenase activity GO: 0005634 nucleus GO: 0005737 cytoplasm GO: 0006096 glycolytic process GO: 0006754 ATP biosynthetic process GO: 0019244 lactate biosynthetic process from pyruvate GO: 0019516 lactate oxidation GO: 0030317 sperm motility GO: 0031514 motile cilium KO: K00016 L-lactate dehydrogenase [EC:1.1.1.27] ReactionKEGG: R00703 (S)-Lactate + NAD+ <=> Pyruvate + NADH + H+ ReactionKEGG: R01000 2-Hydroxybutanoic acid + NAD+ <=> 2-Oxobutanoate + NADH + H+ ReactionKEGG: R03104 3-Mercaptolactate + NAD+ <=> Mercaptopyruvate + NADH + H+ OMIM: 150150 LACTATE DEHYDROGENASE C; LDHC Distance to locus: 112361 LD R2 of rs2403254 with intragenic SNP rs4417246: 0.00024131 Gene: 645312 (GLTPP1) Chr 11 18210451-18212785 (+) 18261224 The intronless GLTP gene at 11p15.1 was transcriptionally silent, primate-specific, and yielded inactive protein. A 5-exon/4-intron GLTP gene at 12q24.11 was the highly conserved, transcript source of active GLTP in human and vertebrate cells. Distance to locus: 129319 LD R2 of rs2403254 with intragenic SNP rs2445180: 0.017123 Gene: 117196 (MRGPRX4) Chr 11 18194384-18195827 (+) 20379614 Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) Protein: MRGX4 (Q96LA9) Mas-related G-protein coupled receptor member X4; GO: 0004930 G-protein coupled receptor activity GO: 0005886 plasma membrane GO: 0016021 integral component of membrane KO: K08396 Mas-related G protein-coupled receptor member X OMIM: 607230 MAS-RELATED G PROTEIN-COUPLED RECEPTOR FAMILY, MEMBER X4; MRGPRX4 Distance to locus: 149345 LD R2 of rs2403254 with intragenic SNP rs2445197: 0.014786 Gene: 645297 (LOC645297) Chr 11 18174824-18175801 (+) Distance to locus: -152228 LD R2 of rs2403254 with intragenic SNP rs11024671: 0.12694 Gene: 160287 (LDHAL6A) Chr 11 18477374-18501147 (+) 18351441 expression of LDHAL6A was able to activate transcriptional activities of AP1(PMA) Pathway: Pyruvate metabolism - Homo sapiens (human) (database: KEGG) Pathway: Glycolysis / Gluconeogenesis - Homo sapiens (human) (database: KEGG) Pathway: Propanoate metabolism - Homo sapiens (human) (database: KEGG) Pathway: Cysteine and methionine metabolism - Homo sapiens (human) (database: KEGG) Pathway: Beta-mercaptolactate-cysteine disulfiduria (database: SMPDB) Pathway: Pyruvate Dehydrogenase Complex Deficiency (database: SMPDB) Pathway: Primary hyperoxaluria II, PH2 (database: SMPDB) Pathway: Pyruvate kinase deficiency (database: SMPDB) Pathway: Gluconeogenesis (database: SMPDB) Pathway: Leigh Syndrome (database: SMPDB) Pathway: Glycogenosis, Type IA. Von gierke disease (database: SMPDB) Pathway: Glycogenosis, Type IC (database: SMPDB) Pathway: Cysteine Metabolism (database: SMPDB) Pathway: Glycogen Storage Disease Type 1A (GSD1A) or Von Gierke Disease (database: SMPDB) Pathway: Pyruvate Metabolism (database: SMPDB) Pathway: Pyruvate Decarboxylase E1 Component Deficiency (PDHE1 Deficiency) (database: SMPDB) Pathway: Triosephosphate isomerase (database: SMPDB) Pathway: Fructose-1,6-diphosphatase deficiency (database: SMPDB) Pathway: Phosphoenolpyruvate carboxykinase deficiency 1 (PEPCK1) (database: SMPDB) Pathway: Glycogenosis, Type IB (database: SMPDB) Pathway: pyruvate fermentation to lactate (database: HumanCyc) Protein: LDH6A (Q6ZMR3) L-lactate dehydrogenase A-like 6A; EC: 1.1.1.27 L-lactate dehydrogenase. (S)-lactate + NAD(+) = pyruvate + NADH. GO: 0004459 L-lactate dehydrogenase activity GO: 0005737 cytoplasm GO: 0006096 glycolytic process KO: K00016 L-lactate dehydrogenase [EC:1.1.1.27] ReactionKEGG: R00703 (S)-Lactate + NAD+ <=> Pyruvate + NADH + H+ ReactionKEGG: R01000 2-Hydroxybutanoic acid + NAD+ <=> 2-Oxobutanoate + NADH + H+ ReactionKEGG: R03104 3-Mercaptolactate + NAD+ <=> Mercaptopyruvate + NADH + H+ Distance to locus: 165119 LD R2 of rs2403254 with intragenic SNP rs4619132: 0.044144 Gene: 117195 (MRGPRX3) Chr 11 18142502-18160027 (+) 15809047 overexpression of the human MrgX3 gene causes a disturbance of the normal cell-differentiation process Protein: MRGX3 (Q96LB0) Mas-related G-protein coupled receptor member X3; GO: 0004930 G-protein coupled receptor activity GO: 0005886 plasma membrane GO: 0016021 integral component of membrane KO: K08396 Mas-related G protein-coupled receptor member X OMIM: 607229 MAS-RELATED G PROTEIN-COUPLED RECEPTOR FAMILY, MEMBER X3; MRGPRX3 Distance to locus: 176712 LD R2 of rs2403254 with intragenic SNP rs11024688: 0.11217 Gene: 7251 (TSG101) Chr 11 18501858-18548503 (-) 23408603 These results support a model in which both HIV-1 Gag-induced membrane curvature and Gag-ESCRT interactions promote tetherin recruitment, but the recruitment level achieved by the former is sufficient for full restriction. 23217182 Data indicate tht n the Biaka, strong signal of selection was detected at CUL5 and at TSG101. 22768867 The expression of TSG101 in HCC is higher than that in corresponding non-cancer tissues and the expression level is closely correlated with TNM stage and metastasis of HCC. 22675076 The results provide evidence for a two-step splicing pathway of the TSG101 mRNA in which the initial constitutive splicing removes all 14 authentic splice sites, thereby bringing the weak alternative splice sites into close proximity. 22348143 identified TSG101 as a novel FIP4-binding protein, which can also bind FIP3. alpha-helical coiled-coil regions of both TSG101 and FIP4 mediate the interaction with the cognate protein 21880841 Depletion of endogenous Tsg101 by siRNA led to a significant reduction of HEV release in cultured cells. 21528537 HIV-1 infection affects the expression of host factors TSG101 and Alix 21455631 Overexpression of PEG10 and TSG101 was detected in gallbladder adenocarcinoma. 21117030 TSG101 knockdown in breast cancer cells induces apoptosis and inhibits proliferation. TSG101 may play a biological role through modulation of the MAPK/ERK signaling pathway in breast cancer. 20504928 Taken together, these data indicate that Marburg virus nucleoprotein enhances budding of virus-like particles by recruiting Tsg101 to the VP40-positive budding site through a PSAP late-domain motif. 20399684 show that ubiquitin recognition by TSG101 is required for cSMAC formation, T cell receptor (TCR) microcluster signal termination, TCR downregulation. 20372822 Results suggest that TSG101 down-regulation in cervical cancer cells is not regulated by genetic or epigenetic events. 19787439 TSG101 may induce the malignant phenotype of cells. 19692479 Data suggest that HSV-1 production is independent of ALIX and TSG101 expression. 19520058 Ca2+-loaded ALG-2 bridges Alix and TSG101 as an adaptor protein. 19362095 molecular dynamics simulations study of the association of the ubiquitin E2 variant domain of the protein Tsg101 and an HIV-derived nonapeptide 19282983 Nucleocapsid region of Gag cooperates with PTAP in the recruitment of cellular proteins necessary for its L domain activity and binds the Bro1-CHMP4 complex required for LYPX(n)L-mediated budding. 19143632 [review] 18600204 Persistent upregulation in colorectal carcinoma cases studied. 18367816 study found that not only the PTAP sequence in the GAT domain but also the PSAP sequence in the C-terminal region of Tom1L1 is responsible for its interaction with the UEV domain of Tsg101 and competes with the HIV-1 Gag protein for the Tsg101 interaction 18077552 Tal polyubiquitinates lysine residues in the C-terminus of uncomplexed Tsg101, resulting in proteasomal degradation. 17940959 Ebola virus can use vacuolar protein surting proteins independently of TSG101 for budding and reveal vacuolar sorting protein 4 as a potential target for filovirus therapeutics. 17853893 that ALIX and TSG101/ESCRT-I also bind a series of proteins involved in cytokinesis, including CEP55, CD2AP, ROCK1, and IQGAP1. 17606716 TSG101 negatively regulates p21 levels, and up-regulation of TSG101 is associated with poor prognosis in ovarian cancer 17556548 study shows that two proteins involved in HIV-1 budding-Tsg101, a subunit of the endosomal sorting complex required for transport I (ESCRT-I), & Alix, an ESCRT-associated protein-were recruited to the midbody during cytokinesis by interaction with Cep55 17428861 ALIX can have a dramatic effect on HIV-1 release by binding at the CHMP4B site; the ability to use ALIX may allow HIV-1 to replicate in cells that express only low levels of Tsg101 17369844 the Tsg101 protein has only weak oncogenic properties 17321722 These data suggest that an intracellular calcium store independent PKC-Sp1 signaling pathway induces early keratinocyte differentiation through upregulation of TSG101. 17229889 TSG101 is a specific Mahogunin substrate 17182691 role for TSG101 in the replication of EBV, a DNA virus, that differs from what is observed for RNA viruses, where TSG101 aids mainly in the endosomal sorting of enveloped late viral proteins for assembly at the plasma membrane 17110434 These results demonstrate that TSG101 is important for CITED2- and HIF-1alpha-mediated cellular regulation in ovarian carcinomas. 17060450 analysis of mechanism of formation of the principal MDM2 isoforms, differential effects of p53 on the production of these isoforms, and differential abilities of human MDM2 isoforms as regulators of the MDM2/TSG101 and p53/MDM2 feedback control loops 17014699 Four proteins (TSG101,Hrs,Aip1/Alix, and Vps4B) of the ESCRT (endosomal sorting complex required for transport) machinery were localized in T cells and macrophages by quantitative electron microscopy. 16707569 These results indicate that Tsg101 is required for the formation of stable vacuolar domains within the early endosome that develop into multivesicular body (MVBs) and Hrs is required for the accumulation of internal vesicles within MVBs. 16552148 The crystal structure of the TSG101 UEV domain (TSG101-UEV) is presented. 16407257 the ORF3 protein exploits the endosomal sorting machinery to enhance the secretion of an immunosuppressant molecule (alpha1 microglobulin) from cultured hepatocytes 16256744 The Tsg101 proteins function in endosomal sorting and are required to incorporate late endosomes into multivesicular bodies. 16138902 Data suggest that RSV and HIV-1 Gag direct particle release through independent ESCRT-mediated pathways that are linked through Tsg101-Nedd4 interaction. 16004603 The expression and transport of ALG-2 in association with TSG101 and Vps4B are reported. 15908698 interaction of Gag with Tsg101 and Alix favors budding from the plasma membrane and relieves a requirement for ubiquitination by Nedd4 15657031 TSG101 binds GR and protects the non-phosphorylated receptor from degradation. 15126635 Tsg101 and Nedd4.1 act successively in the assembly process of HTLV-1 to ensure proper Gag trafficking through the endocytic pathway up to late endosomes where the late steps of retroviral release occur. 15053872 X-ray crystallography study of the UEV domain of TSG101 and ubquitin showed the basis for the binding recognition at high resolution. 15033475 molecular interactions between Daxx and TSG101 establish an efficient repressive transcription complex in the nucleus 14991575 Reduction of TSG101 protein has a negative impact on breast and prostate tumor cell growth 14761944 TSG101 activates androgen receptor-induced transcription by transient stabilization of the monoubiquitinated state 14526201 alternative splicing and role implicated in interaction with HIV-1 12802020 the TSG101 interaction with HRS is a crucial step in endocytic down-regulation of mitogenic signaling and this interaction may have a role in linking the functions of early and late endosomes 12743307 truncated and full length forms of TSG101 inhibit HIV-1 budding by interacting with the p6 L domain and by disrupting the cellular endosomal sorting machinery 12725919 Human ortholog TSG101 does not substitute VPS23 in its ability to rescue the phenotype of defective plasma membrane proteins 12505256 Analysis of BRCA1, TP53, and TSG101 germline mutations in German breast and/or ovarian cancer families. 12388682 interacts specifically with human immunodeficiency virus type 2 gag polyprotein, results in increased levels of ubiquinated gag, and is incorporated into HIV-2 virions 12379843 solution structure of the UEV (ubiquitin E2 variant) binding domain of Tsg101 in complex with a PTAP peptide that spans the late domain of HIV-1 p6(Gag) 12006492 structure and functional interactions of its binding sites 11943869 Negative regulation of cell growth and differentiation by TSG101 through association with p21(Cip1/WAF1). 11916981 recognize ubiquitin and act in the removal of endosomal protein-ubiquitin conjugates. 11838966 TSG101 expression in gynecological tumors: relationship to cyclin D1, cyclin E, p53 and p16 proteins. Pathway: Endocytosis - Homo sapiens (human) (database: KEGG) Pathway: Internalization of ErbB1 (database: PID) Pathway: HIV Life Cycle (database: Reactome) Pathway: HIV Infection (database: Reactome) Pathway: Disease (database: Reactome) Pathway: TCR (database: NetPath) Pathway: Late Phase of HIV Life Cycle (database: Reactome) Pathway: Budding and maturation of HIV virion (database: Reactome) Pathway: Membrane binding and targetting of GAG proteins (database: Reactome) Pathway: Endosomal Sorting Complex Required For Transport (ESCRT) (database: Reactome) Pathway: Membrane Trafficking (database: Reactome) Protein: TS101 (Q99816) Tumor susceptibility gene 101 protein; GO: 0000813 ESCRT I complex GO: 0001558 regulation of cell growth GO: 0003677 DNA binding GO: 0003714 transcription corepressor activity GO: 0005515 protein binding GO: 0005634 nucleus GO: 0005737 cytoplasm GO: 0005769 early endosome GO: 0005770 late endosome GO: 0005771 multivesicular body GO: 0006464 cellular protein modification process GO: 0007050 cell cycle arrest GO: 0008285 negative regulation of cell proliferation GO: 0010008 endosome membrane GO: 0015031 protein transport GO: 0016032 viral process GO: 0016197 endosomal transport GO: 0019058 viral life cycle GO: 0019068 virion assembly GO: 0019082 viral protein processing GO: 0030216 keratinocyte differentiation GO: 0031625 ubiquitin protein ligase binding GO: 0031902 late endosome membrane GO: 0043130 ubiquitin binding GO: 0043162 ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway GO: 0045892 negative regulation of transcription, DNA-templated GO: 0046755 viral budding GO: 0048306 calcium-dependent protein binding GO: 0051301 cell division GO: 0061024 membrane organization GO: 0070062 extracellular vesicular exosome GO: 0075733 intracellular transport of virus KO: K12183 ESCRT-I complex subunit TSG101 OMIM: 601387 TUMOR SUSCEPTIBILITY GENE 101; TSG101 OMIM: 114480 BREAST CANCER Distance to locus: 186640 Gene: 100422666 (LOC100422666) Chr 11 18511786-18512501 (-) Distance to locus: -187467 LD R2 of rs2403254 with intragenic SNP rs2468782: 0.044144 Gene: 6290 (SAA3P) Chr 11 18134019-18137679 (-) Pathway: Selenium Pathway (database: Wikipathways) Pathway: Vitamin B12 Metabolism (database: Wikipathways) Pathway: Folate Metabolism (database: Wikipathways) Distance to locus: 196753 LD R2 of rs2403254 with intragenic SNP rs871699: 0.048632 Gene: 100289300 (HIGD1AP5) Chr 11 18122973-18128393 (+) Distance to locus: -197508 LD R2 of rs2403254 with intragenic SNP rs3741199: 0.053612 Gene: 113174 (SAAL1) Chr 11 18101890-18127638 (-) 22127701 Overexpression of SPACIA1/SAAL1, a newly identified gene that is involved in synoviocyte proliferation, accelerates the progression of synovitis in mice and humans. Protein: SAAL1 (Q96ER3) Protein SAAL1; GO: 0005576 extracellular region GO: 0006953 acute-phase response Distance to locus: 228098 LD R2 of rs2403254 with intragenic SNP rs7119085: 0.11619 Gene: 55293 (UEVLD) Chr 11 18553244-18610281 (-) 12427560 identification, sequencing and analysis of cDNA in various colon carcinoma cell lines as well as normal and tumor samples from colon Protein: UEVLD (Q8IX04) Ubiquitin-conjugating enzyme E2 variant 3; GO: 0006464 cellular protein modification process GO: 0015031 protein transport GO: 0016616 oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor GO: 0044262 cellular carbohydrate metabolic process GO: 0070062 extracellular vesicular exosome OMIM: 610985 UBIQUITIN E2 VARIANT AND LACTATE/MALATE DEHYDROGENASE DOMAINS-CONTAINING PROTEIN; UEVLD Distance to locus: -262811 LD R2 of rs2403254 with intragenic SNP rs2056246: 0.04383 Gene: 7166 (TPH1) Chr 11 18042084-18062335 (-) 23597148 Our results suggest that in acute depression TPH1 A218C polymorphism and specifically the CC genotype together with the information on remission or treatment response differentiates between different temperament profiles and their changes. 23328530 TPH1 deficiency or inhibition reduces allergic airway inflammation. Platelet-derived 5-HT is pivotal in AAI and lack of 5-HT leads to impaired Th2 priming capacity of bone marrow dendritic cells. 23177301 There is a relationship between sex, age & the TPH1 locus, with a trend towards a lower frequency of the AA genotype in former smokers. The TPH1 polymorphism is an indicator of therapeutic failure in smoking cessation. 23157339 This study detected allelic or genotypic associations of TPH1 in clinically significant depression in Alzheimer;s disease . 23088179 Associations between adolescents' physical activity and depressive symptoms are not modified by plasticity genes. 22781862 Among maltreated children, polymorphisms of TPH1 were related to heightened self-report and peer report of antisocial behavior. 22697203 Aggression in MDD patients is more susceptible to an excess of TPH1 CC homozygote than in undifferentiated somatoform disorder patients. TPH1 gene is most likely to have a shared effect on aggression and MDD. 22483952 variants in TPH1 gene constitute risk factors for post traumatic stress disorder symptoms. 22429480 The GGCCGGGC haplotype in the first haplotype block of TPH1 was significantly associated with middle insomnia. 22323131 In conclusion, regardless of visceral hypersensitivity state, several serotonergic signaling components are altered in IBS patients. 22053918 results indicate that the TPH1 CC recessive genotype is likely to be a genetic risk factor for criminal behavior, especially homicidal behavior in patients with schizophrenia. 21989108 Variation in TPH1 may increase risk for developing borderline personality disorder as a result of childhood abuse. 21836641 In non-diabetic controls, SNPs of TPH1 were associated with waist circumference and BMI. 21601290 The TPH1 A218C polymorphism is a potential biomarker for bipolar disorder and alcohol dependence risk in Caucasian population (Meta-Analtsis) 21308753 we concluded that TRH1 SNP is not associated with either adolescent idiopathic scoliosis predisposition or curve severity in Japanese. 21073637 Results describe associations of tryptophan hydroxylase 1 and 2 gene variants with irritable bowel syndrome-related GI symptoms and stool characteristics. 20945066 Clinical trial of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 20945066 This study indicated that the TPH1 218A/C genotype and allele frequencies differed between Taiwanese healthy controls and patients with major depressive disorder 20921119 Family analysis of 38 TPH1 mutation carriers and 41 of their offspring revealed that offspring of mothers carrying TPH1 mutations reported 1.5- to 2.5-times-higher ADHD scores and related symptoms during childhood and as adults than did controls. 20580984 The results suggest that TPH1 gene variation participates in the regulation of serotonin and dopamine turnover rates in the central nervous system of healthy human subjects. 20213726 In the single largest attention-deficit/hyperactivity disorder (ADHD) genetic study of TPH1 and TPH2 variants presented to date (n = 3,559 individuals), evidence for a substantial effect of common genetic variants on persistent ADHD, was not found. 20213726 Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 20198979 Association between the tryptophan hydroxylase (TPH) gene polymorphic markers and endogenous psychoses 20144688 Neither common genetic variations of TPH1 nor TPH2 are likely to contribute to the genetic susceptibility to schizophrenia in Japanese population. 20046510 The findings suggest that TPH1 does not play a major role in aggressive behavior via anger in schizophrenic patients. 19961731 The TPH gene A218C polymorphism is not associated with essential hypertension in Chinese northern Han population. 19939457 Studies indicate that the mechanism of oxygen activation in tryptophan hydroxylase does not require inversion of spin. 19874868 In this Finnish population TPH1 218A/C polymorphism was associated with the risk of MDD and treatment response; CC genotype was associated with the increased risk of MDD and lower probability of responding treatment. 19816759 The catalytic domains of human tryptophan hydroxylase 1 and 2 have been expressed, purified and the kinetic properties have been studied and are compared. 19744316 Expression of DKK1 and TPH1 were negatively regulated by ASCL1 in pancreatic endocrine tumours. 19590397 evaluation of association of 7 serotonin signal transduction-linked SNP's [HTR1A (rs6295), HTR2A (rs6313, rs6311 & rs7997012), HTR6 (rs1805054), TPH1 (rs1800532) & TPH2 (rs1386494)] with major depressive disorder; no association found 19526457 Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 19526457 tryptophan hydroxylase 1 (TPH1) has a role in schizophrenia susceptibility and suicidal behavior 19506906 Meta-analysis of gene-disease association. (HuGE Navigator) 19500158 Our results suggest that the A218C polymorphism of the TPH1 gene serves as a modulator of amygdala activity in patients with MDD. 19474754 we found significant associations of bodyweight gain by more than 4 kg during antidepressant treatment with polymorphisms in genes coding for COMT and TPH1. 19445671 TPH1 were expressed in all brain regions similarly except for within the striatum and cerebellum, where TPH1 was expressed at a significantly higher level than TPH2. 19381154 TPH1 variation (rs10488683) was relevant in the diathesis for suicide attempts. 19233335 neuronal TPH1 & TPH2 expression in anterior pituitary; evidence against a strictly separated duality of serotonergic system; TPH1 may also have impact on neuronal mechanisms via hypothalamic-pituitary-adrenal axis regulation by its pituitary localization 19220488 TPH-1 genotype could confer a vulnerability to suicidal behaviour through a reduced capacity to control anger 19181488 In human no significant TPH1 expression was detected in the brain. 19095219 Clinical trial of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19077664 Clinical trial of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 19052197 Hence, our study shows, for the first time, evidence of a link between genetically controlled serotonergic modulation of amygdala activity and placebo-induced anxiety relief. 19032713 Meta-analysis of gene-disease association. (HuGE Navigator) 18982004 Meta-analysis of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) 18937498 the chicken TPH1.Trp.imidazole structure resembles the PAH.BH 4.thienylalanine structure more (root-mean-square deviation for Calpha atoms of 0.90 A) than the human TPH1 structure (root-mean-square deviation of 1.47 A). 18826425 TPH-1 haplotype displayed a significantly higher frequency in patients with borderline personality disorder with impaired decision making 18813430 The effect of TPH gene polymorphism on TMD development and whether AA genotype or A allele are preventive factors for TMD remains unclear. 18794762 These results suggested that TPH1 was an adolescent idiopathic scoliosis predisposition gene. 18583979 Meta-analysis of gene-disease association. (HuGE Navigator) 18562131 792 cigarette smokers from the Patch in Practice trial were genotyped for the tryptophan hydroxylase (TPH1 A779C), serotonin transporter (SLC6A45-HTTLPR), and 5-HT1A (HTR1A C-1019G) polymorphisms. 18562131 Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 18506706 Results suggest that the A allele of the tryptophan hydroxylase-1 A218 polymorphism may be associated with Borderline Personality Disorder, and that it does not appear to be related to suicidal behavior in this population. 18402117 Polymorphism of the TPH gene-T457C locus could show ethnic and regional differences. 18366104 Three polymorphisms with theoretical and/or empirical ties to ASB or related traits (i.e., tryptophan hydroxylase-A218C, 5HT(2A) His452Tyr, and the DAT1 variable nucleotide tandem repeat) were genotyped. 18332644 The result suggests that 218A/C variants of TPH1 cannot play a major role as predictor of treatment response as well as intolerance in Japanese patients with major depression. 18221792 It is suggested that TPH1 protein may be implicated in schizophrenia with beta-thalassemia simultaneously, present on the short arm of chromosome 11. 18181017 Allele frequencies of three TPH2 variants and a TPH1 variant vary significantly among four ethnic groups with heroid adddiction. 18177948 TPH1 gene is implicated in depression. 17986837 TPH1 779A/C SNP is related to nicotine dependence and is associated with smoking status discriminating smokers from never-smokers and showed a tendency for an association with the degree of nicotine dependence. 17870198 Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 17870198 There were no significant associations between the polymorphisms or haplotypes of TPH1 and schizophrenia in our Japanese subjects. Our updated meta-analysis suggests the possible involvement of the TPH1 218A allele in schizophrenia. 17548152 These findings found that tryptophan-hydroxylase-1 A218C polymorphism does not play a part in the genetic susceptibility to bulimia nervosa, but to be involved in predisposing bulimic patients to a more disturbed eating behavior and higher harm avoidance. 17521439 TPH1 A779C C/A genotype was inversely associated with good treatment response to neuroleptics when compared with non-responding patients with schzophrenia 17509016 Results describe regional differences in expression of TPH-1, SERT, 5-HT(3) and 5-HT(4) receptors in the stomach and duodenum. 17221847 study reports the learning process of decision-making in suicide attempters to be modulated by four serotonergic gene polymorphisms, 5HTTLPR, TPH1 A218C, MAOA u-VNTR, and TPH2 rs1118997 17194593 Since it is unlikely that TPH polymorphism alters serotonin biosynthesis, its association with migraine may be attributed to linkage disequilibrium with a functional variant within the TPH gene or a nearby gene. 17066254 We studied the association between tryptophan hydroxylase 1 (TPH1) A218C polymorphism and treatment response in electroconvulsive therapy (ECT). 17026953 These findings do not support a major role for common 5-hydroxytryptamine transporter, TPH1, and monoamine oxidase A polymorphisms in contributing to susceptibility to premenstrual dysphoric disorder. 16958027 It appears unlikely that the TPH1 and TPH2 genes play a significant role in the susceptibility to autism or to autism endophenotypes including severe obsessive-compulsive behaviors and self-stimulatory behaviors. 16848783 The presence of the A/A haplotype of the TPH1 intron 7 polymorphism predicted a high level of adulthood harm avoidance in the presence of a hostile childhood environment. Findings also suggest a gene-environment correlation for novelty seeking in men. 16822601 polymorphisms play no significant roles in the pathogenesis and clinical symptomatologiy of panic dissorder in a Korean population. 16806098 Single marker association analyses showed two SNPs significantly associated with schizophrenia. A "sliding window" analysis attributed the strongest disease association to a haplotype configuration localized between the promoter region and intron 3. 16741719 Meta-analysis of gene-disease association. (HuGE Navigator) 16716203 A218C polymorphism of the TPH gene may not be a susceptibility factor for suicidal behavior in this group of psychiatric patients but confirm that a family suicidal behavior history increases the proband's suicide attempt risk. 16538180 The A218C SNP in TPH1 is not associated with completed suicide. 16467214 TPH1 may play a significant role in the aetiology of psychiatric disorders in the Han Chinese population. 16450114 Meta-analysis of gene-disease association. (HuGE Navigator) 16450114 Strong overall association between suicidal behavior and the A779C/A218C polymorphisms, supporting the involvement of TPH in the pathogenesis of suicidal behavior. 16389593 Haplotype analyses showed that the rare 218A/-6526G haplotype was significantly not transmitted to probands with Attention deficit hyperactivity disorder. 16389591 The TPH CC genotype in elderly victims of violent suicide points to the possible combined effect of the respective genetic factor and physiological changes during aging on the predisposition to this disorder. 16314762 A218C polymorphism of the TPH gene does not play a major role in pathogenesis in major depressive disorder and does not serve as a modulator of antidepressant activity. 16165107 te most common TPH-1 variants appear to carry no risk, while some of the less frequent variants might contribute to genetic predisposition to major depression 15799788 Single nucleotide polymorphisms in the tryptophan hydroxylase gene are not associated with bipolar affective disorder. 15722951 Tryptophan hydroxylasse polymorphisms not statistically diffrent in general anxiety disorder in Chinese. 15654285 Polymorphisms in the promoter region may influence the function of the TPH1 gene and further influence the proclivity of alcohol dependence in one ethnic group in Taiwan. 15635702 The positive heterosis effects with respect to nicotine addiction and personality support the idea that the TPH1 gene exerts pleiotropic effects. 15544576 Five single nucleoside polymorphisms of the TPH1 gene were studied in postpartum women with depression or/and anxiety. Genotype frequency differences for the T27224C polymorphism were found between the comorbid and normal groups. 15211625 There was a nearly doubling of the risk for childhood-onset schizophrenia associated with the AA genotype compared to other genotype groups in a Japanese sample 15182943 This study suggested that aggression in male subjects with Alzheimer's disease may be genetically linked to polymorphic variation at the tryptophan hydroxylase gene. 15135070 the denatured state properties of the AAAHs (TH, TPH and PAH) contribute significantly to the stability of these enzymes and their tolerance towards missense mutations 15052272 Clinical trial of gene-environment interaction and pharmacogenomic / toxicogenomic. (HuGE Navigator) 15052272 Study assessed genetic factors influencing antidepressant response to fluoxetine. Results implicate TPH1 in general response to fluoxetine. 14681922 Meta-analysis of gene-disease association. (HuGE Navigator) 14504413 Population-based association study tested the hypothesis that the tryptophan hydroxylase (TPH) A218C was associated with TPQ personality trait scores in a sample population of 209 young healthy Chinese. 12960746 Association of tryptophan hydroxylase gene polymorphisms and unipolar depression in a case-control study design. 12915291 Observational study and meta-analysis of gene-disease association. (HuGE Navigator) 12476329 THis enzyme's immunoreactivity is altered by the genetic variation in postmortem brain samples of both suicide victims and controls. 12379098 X-ray crystal structure analysis of a truncated functional form of TPH with the bound cofactor analogue 7,8-dihydro-L-biopterin provides the first atomic-resolution information for the catalytic domain and cofactor binding site of the enzyme. 12208565 Clinical trial of gene-environment interaction. (HuGE Navigator) 12116191 Meta-analysis of gene-disease association. (HuGE Navigator) 11992558 study did not support the involvement of 5-HTTLPR, TPH, MAO-A, or DRD4 polymorphisms in mood disorders 11747434 In a stable N-terminally truncated form of TPH that includes the catalytic domain, residue Phe-313 interacts with the substrate through ring stacking, thus predicting substrate specificity. 11597824 Clinical trial of gene-environment interaction. (HuGE Navigator) 11526473 Clinical trial of gene-disease association, gene-gene interaction, and gene-environment interaction. (HuGE Navigator) Pathway: Tryptophan metabolism - Homo sapiens (human) (database: KEGG) Pathway: Serotonergic synapse - Homo sapiens (human) (database: KEGG) Pathway: Selective Serotonin Reuptake Inhibitor Pathway, Pharmacodynamics (database: PharmGKB) Pathway: Tryptophan Metabolism (database: SMPDB) Pathway: Tryptophan metabolism (database: Wikipathways) Pathway: Biogenic Amine Synthesis (database: Wikipathways) Pathway: SIDS Susceptibility Pathways (database: Wikipathways) Pathway: Metabolism of amino acids and derivatives (database: Reactome) Pathway: Serotonin and melatonin biosynthesis (database: Reactome) Pathway: serotonin and melatonin biosynthesis (database: HumanCyc) Pathway: Tryptophan degradation (database: INOH) Pathway: tryptophan utilization I (database: HumanCyc) Pathway: tryptophan utilization II (database: HumanCyc) Pathway: Amine-derived hormones (database: Reactome) Protein: TPH1 (P17752) Tryptophan 5-hydroxylase 1; EC: 1.14.16.4 Tryptophan 5-monooxygenase. L-tryptophan + tetrahydrobiopterin + O(2) = 5-hydroxy-L-tryptophan + 4a-hydroxytetrahydrobiopterin. GO: 0004510 tryptophan 5-monooxygenase activity GO: 0005506 iron ion binding GO: 0005829 cytosol GO: 0007623 circadian rhythm GO: 0009072 aromatic amino acid family metabolic process GO: 0016597 amino acid binding GO: 0030279 negative regulation of ossification GO: 0034641 cellular nitrogen compound metabolic process GO: 0035902 response to immobilization stress GO: 0042427 serotonin biosynthetic process GO: 0043005 neuron projection GO: 0044281 small molecule metabolic process GO: 0046219 indolalkylamine biosynthetic process GO: 0046849 bone remodeling GO: 0060749 mammary gland alveolus development KO: K00502 tryptophan 5-monooxygenase [EC:1.14.16.4] ReactionKEGG: R01814 Tetrahydrobiopterin + L-Tryptophan + Oxygen <=> 5-Hydroxy-L-tryptophan + Dihydrobiopterin + H2O ReactionKEGG: R07213 L-Tryptophan + Tetrahydrobiopterin + Oxygen <=> 5-Hydroxy-L-tryptophan + 4a-Hydroxytetrahydrobiopterin OMIM: 191060 TRYPTOPHAN HYDROXYLASE 1; TPH1 Distance to locus: -290509 LD R2 of rs2403254 with intragenic SNP rs4402275: 0.097315 Gene: 26297 (SERGEF) Chr 11 17809595-18034637 (-) 12459492 DelGEF binds to the human homologue of Sec5 and modulates secretion. Protein: SRGEF (Q9UGK8) Secretion-regulating guanine nucleotide exchange factor; GO: 0005087 Ran guanyl-nucleotide exchange factor activity GO: 0005515 protein binding GO: 0005634 nucleus GO: 0005737 cytoplasm GO: 0007165 signal transduction GO: 0032316 regulation of Ran GTPase activity GO: 0050709 negative regulation of protein secretion KO: K15421 secretion-regulating guanine nucleotide exchange factor OMIM: 606051 SECRETION-REGULATING GUANINE NUCLEOTIDE EXCHANGE FACTOR; SERGEF Distance to locus: 291855 LD R2 of rs2403254 with intragenic SNP rs6416020: 0.10506 Gene: 100129966 (LOC100129966) Chr 11 18617001-18617913 (-) Distance to locus: -296205 LD R2 of rs2403254 with intragenic SNP rs16935596: 0.1049 Gene: 100506540 (LOC100506540) Chr 11 18621351-18631802 (+) Distance to locus: 302802 Gene: 144108 (SPTY2D1) LD R2 of rs2403254 with intragenic SNP rs16935596: 0.1049 Chr 11 18627948-18656020 (-) Protein: SPT2 (Q68D10) Protein SPT2 homolog; KO: K15193 protein SPT2 Distance to locus: -361331 LD R2 of rs2403254 with intragenic SNP rs11024754: 0.092861 Gene: 100506569 (LOC100506569) Chr 11 18686477-18688031 (+) Distance to locus: -395205 LD R2 of rs2403254 with intragenic SNP rs14792: 0.024249 Gene: 144110 (TMEM86A) Chr 11 18720351-18726332 (+) Protein: TM86A (Q8N2M4) Lysoplasmalogenase-like protein TMEM86A; GO: 0016021 integral component of membrane Distance to locus: 400706 LD R2 of rs2403254 with intragenic SNP rs1867877: 0.035306 Gene: 283284 (IGSF22) Chr 11 18725852-18747777 (-) Protein: IGS22 (Q8N9C0) Immunoglobulin superfamily member 22; Distance to locus: 424329 LD R2 of rs2403254 with intragenic SNP rs7950091: 0.10383 Gene: 84867 (PTPN5) Chr 11 18749475-18813389 (-) 22885232 This experiments demonstrated that deletion of STEP can enhance experience-induced neuroplasticity and memory formation 22649233 This study identified a novel role for PTPN5 in mediating the development of stress-related cognitive and morphological changes. 22555153 The results imply a model in which PTPN5 may play a role in normal cognitive functioning and contribute to aspects of the neuropathology of schizophrenia. 21883219 STEP(61kDa) is required for Abeta transgene-mediated internalization of GluA1/GluA2 glutamate receptors in a transgenic mousemodel. 20699650 STEP contributes to aspects of the pathophysiology in Alzheimer's disease; loss of GluN1/GluN2B subunits from neuronal membranes and Abeta-mediated NMDAR internalization are discussed 20427654 findings show that STEP(61) levels are progressively increased in the prefrontal cortex of Alzheimer disease brains 20379614 Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) 19000305 in colorectal tumors, microsatellite repeats mutation rates are higher than the mean mutation frequency 16441242 determined high-resolution structures of all of the human family members of Mitogen-Activated Protein Kinase-specific protein tyrosine phosphatases Pathway: MAPK signaling pathway - Homo sapiens (human) (database: KEGG) Pathway: MAPK Signaling Pathway (database: Wikipathways) Pathway: EGF-EGFR Signaling Pathway (database: Wikipathways) Protein: PTN5 (P54829) Tyrosine-protein phosphatase non-receptor type 5; EC: 3.1.3.48 Protein-tyrosine-phosphatase. Protein tyrosine phosphate + H(2)O = protein tyrosine + phosphate. GO: 0001784 phosphotyrosine binding GO: 0004725 protein tyrosine phosphatase activity GO: 0005789 endoplasmic reticulum membrane GO: 0006470 protein dephosphorylation GO: 0016021 integral component of membrane KO: K04458 protein-tyrosine phosphatase [EC:3.1.3.48] ReactionKEGG: R02585 Protein tyrosine phosphate + H2O <=> Protein tyrosine + Orthophosphate KO: K18018 tyrosine-protein phosphatase non-receptor type 5 [EC:3.1.3.48] ReactionKEGG: R02585 Protein tyrosine phosphate + H2O <=> Protein tyrosine + Orthophosphate OMIM: 176879 PROTEIN-TYROSINE PHOSPHATASE, NONRECEPTOR-TYPE, 5; PTPN5 footnotes: - Linkage disequilibrium R2 is based on founders in HapMap r27 CEU population