* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Inquiry into Life Twelfth Edition
Short interspersed nuclear elements (SINEs) wikipedia , lookup
DNA vaccination wikipedia , lookup
Cell-free fetal DNA wikipedia , lookup
DNA damage theory of aging wikipedia , lookup
Genome evolution wikipedia , lookup
Molecular cloning wikipedia , lookup
Epigenetics of cocaine addiction wikipedia , lookup
Genome (book) wikipedia , lookup
Gene expression profiling wikipedia , lookup
Neocentromere wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Biology and consumer behaviour wikipedia , lookup
Genomic imprinting wikipedia , lookup
Deoxyribozyme wikipedia , lookup
DNA supercoil wikipedia , lookup
Nucleic acid double helix wikipedia , lookup
Minimal genome wikipedia , lookup
Extrachromosomal DNA wikipedia , lookup
Point mutation wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Epigenetics of depression wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Designer baby wikipedia , lookup
Microevolution wikipedia , lookup
Ridge (biology) wikipedia , lookup
Helitron (biology) wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
History of genetic engineering wikipedia , lookup
Non-coding DNA wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Transcription factor wikipedia , lookup
Epigenetics in stem-cell differentiation wikipedia , lookup
Epigenetics wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Cancer epigenetics wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Primary transcript wikipedia , lookup
Epigenomics wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Epigenetics in learning and memory wikipedia , lookup
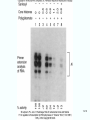
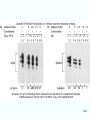
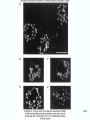
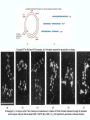
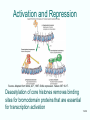
Molecular Biology Lecture 20 Chapter 13 Chromatin Structure and Its Effects on Transcription Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Histones • Eukaryotic cells contain 5 kinds of histones – – – – – H1 H2A H2B H3 H4 Source: Panyim and Chalkley, Arch. Biochem. 13-2 & Biophys. 130, 1969, f. 6A, p.343. Properties of Histones • Pronounced positive charge at neutral pH • Most are well-conserved from one species to another • Not single copy genes, repeated many times – Some copies are identical – Others are quite different – H4 has only had 2 variants ever reported 13-3 Nucleosomes • First order of folding is the nucleosome – X-ray diffraction has shown strong repeats of structure at 100Å intervals – This spacing approximates the nucleosome spaced at 110Å intervals 13-4 Histones in the Nucleosome • Chemical cross-linking in solution: – H3 to H4 – H2A to H2B • H3 and H4 exist as a tetramer (H3-H4)2 • Chromatin is composed of roughly equal masses of DNA and histones – Corresponds to 1 histone octamer per 200 bp of DNA – Octamer composed of: • 2 each H2A, H2B, H3, H4 • 1 each H1 13-5 Fig. 13.4 13-6 H1 and Chromatin • Treatment of chromatin with trypsin or high salt buffer removes histone H1 • This treatment leaves chromatin looking like “beads-on-a-string” • The beads named nucleosomes – Core histones form a ball with DNA wrapped around the outside – H1 also lies on the outside of the nucleosome 13-7 Nucleosome Structure • Central (H3-H4)2 core attached to H2AH2B dimers • Grooves on surface define a left-hand helical ramp – a path for DNA winding – DNA winds almost twice around the histone core condensing DNA length by 6- to 7-X – Core histones contain a histone fold: • 3 a-helices linked by 2 loops • Extended tail of about 28% of core histone mass • Tails are unstructured 13-8 The 30-nm Fiber • Second order of chromatin folding produces a fiber 30 nm in diameter – The string of nucleosomes condenses to form the 30-nm fiber in a solution of increasing ionic strength – This condensation results in another six- to seven-fold condensation of the nucleosome itself • Four nucleosomes condensing into the 30nm fiber form a zig-zag structure 13-9 13-10 Formation of the 30-nm Fiber • Two stacks of nucleosomes form a lefthanded helix – Two helices of polynucleosomes – Zig-zags of linker DNA • Role of histone H1? – 30-nm fiber can’t form without H1 – H1 crosslinks to other H1 more often than to core histones 13-11 13-12 13-13 Higher Order Chromatin Folding • 30-nm fibers account for most of chromatin in a typical interphase nucleus • Further folding is required in structures such as the mitotic chromosomes • Model favored for such higher order folding is a series of radial loops Source: Adapted from Marsden, M.P.F. and U.K. Laemmli, Metaphase chromosome structure: Evidence of a radial loop model. Cell 17:856, 13-14 1979. Chromatin Structure and Gene Activity • Histones, especially H1, have a repressive effect on gene activity in vitro • Two families of 5S rRNA genes studied are oocyte and somatic genes – Oocyte genes are expressed only in oocytes – Somatic genes are expressed both in oocytes and somatic cells – Somatic genes form more stable complexes with transcription factors 13-15 Transcription Factors and Histones Control the 5S rRNA • Genes active by TFIIIs preventing formation of nucleosome stable complexes with internal control region • Stable complexes require histone H1 and exclude TFIIIs once formed so that genes are repressed 13-16 Effects of Histones on Transcription of Class II Genes • Core histones assemble nucleosome cores on naked DNA • Transcription of reconstituted chromatin with an average of 1 nucleosome / 200 bp DNA exhibits 75% repression relative to naked DNA • Remaining 25% is due to promoter sites not covered by nucleosome cores 13-17 13-18 Histone H1 and Transcription • Histone H1 causes further repression of template activity, in addition to that of core histones • H1 repression can be counteracted by transcription factors • Sp1 and GAL4 act as both: – Antirepressors preventing histone repressions – Transcription activators • GAGA factor: – Binds to GA-rich sequences in the Krüppel promoter – An antirepressor – preventing repression by histones 13-19 13-20 Model of Transcriptional Activation Source: Adapted from Laybourn, P.J. and J. T. Kadonaga, Role of nucleosomal cores and histone H1 in regulation of transcription by polymerase II. Science 254:243, 1991. 13-21 Nucleosome Positioning • Model of activation and antirepression asserts that transcription factors can cause antirepression by: – Removing nucleosomes that obscure the promoter – Preventing initial nucleosome binding to the promoter 13-22 Nucleosome-Free Zones • Nucleosome positioning would result in nucleosome-free zones in the control regions of active genes • Assessment in a circular chromosome can be difficult without some type of marker 13-23 13-24 13-25 Detecting DNaseHypersensitive Regions • Active genes tend to have DNase-hypersensitive control regions • Part of this hypersensitivity is due to absence of nucleosomes 13-26 13-27 13-28 Histone Acetylation • Histone acetylation occurs in both cytoplasm and nucleus • Cytoplasmic acetylation carried out by HAT B (histone acetyltransferase, HAT) – Prepares histones for incorporation into nucleosomes – Acetyl groups later removed in nucleus • Nuclear acetylation of core histone N-terminal tails – Catalyzed by HAT A – Correlates with transcription activation – Coactivators of HAT A found which may allow loosening of association between nucleosomes and gene’s control region – Attracts bromodomain proteins, essential for transcription 13-29 Histone Deacetylation • Transcription repressors bind to DNA sites and interact with corepressors which in turn bind to histone deacetylases – Repressors • Unliganded nuclear receptors • Mad-Max – Corepressors • NCoR/SMRT • SIN3 – Histone deacetylases - HDAC1 and 2 13-30 Ternary Protein Complexes • Assembly of complex brings histone deacetylases close to nucleosomes • Deacetylation of core histones allows – Histone basic tails to bind strongly to DNA, histones in neighboring nucleosomes – This inhibits transcription 13-31 Activation and Repression Source: Adapted from Wolfe, A.P., 1997. Sinful repression. Nature 387:16-17. Deacetylation of core histones removes binding sites for bromodomain proteins that are essential for transcription activation 13-32