* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Chromatin Structure and Its Effects on Transcription
Gel electrophoresis of nucleic acids wikipedia , lookup
Transgenerational epigenetic inheritance wikipedia , lookup
Genealogical DNA test wikipedia , lookup
DNA damage theory of aging wikipedia , lookup
United Kingdom National DNA Database wikipedia , lookup
Behavioral epigenetics wikipedia , lookup
Epitranscriptome wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Epigenetics of cocaine addiction wikipedia , lookup
Cell-free fetal DNA wikipedia , lookup
DNA vaccination wikipedia , lookup
Molecular cloning wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Bisulfite sequencing wikipedia , lookup
Microevolution wikipedia , lookup
Point mutation wikipedia , lookup
Designer baby wikipedia , lookup
DNA supercoil wikipedia , lookup
Extrachromosomal DNA wikipedia , lookup
Nucleic acid double helix wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Non-coding DNA wikipedia , lookup
History of genetic engineering wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Epigenetics of depression wikipedia , lookup
Neocentromere wikipedia , lookup
Helitron (biology) wikipedia , lookup
Transcription factor wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Cancer epigenetics wikipedia , lookup
Epigenetics wikipedia , lookup
Epigenetics in stem-cell differentiation wikipedia , lookup
Primary transcript wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Epigenomics wikipedia , lookup
Epigenetics in learning and memory wikipedia , lookup
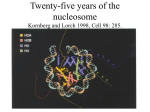
Chromatin Structure and Its Effects on Transcription Chapter 13 Histones • Eukaryotic cells contain 5 kinds of histones – – – – – H1 H2A H2B H3 H4 • Each histone type - not homogenous – Gene reiteration – Posttranslational modification Features of Histones • Abundant proteins • Most are well-conserved from one species to another • Not single copy genes - repeated many times – Some copies are identical – Others are quite different – H4 has - 2 variants Nucleosomes • Chromosomes are long, thin molecules –tangle - if not carefully folded • Folding occurs in several ways - First order of folding is the nucleosome – Maurice Wilkins - X-ray diffraction has shown strong repeats of structure at 100Å intervals – 110Å spacing Histones in the Nucleosome • Chemical cross-linking in solution: (Korenberg) – H3 to H4 – H2A to H2B • H3 and H4 exist as a tetramer (H3-H4)2 • Chromatin is composed of roughly equal masses of DNA and histones – 1 histone octamer/200 bp of DNA – Octamer composed of: • 2 each of H2A, H2B, H3, H4 • 1 each of H1 H1 and Nucleosomes • Treatment of chromatin with trypsin or high salt buffer removes histone H1 • Left chromatin looking like “beads-on-a-string” • The beads named nucleosomes – Core histones form a ball with DNA wrapped around the outside – H1 also lies on the outside of the nucleosome Histone Acetylation • Histone acetylation occurs in both cytoplasm and nucleus • Cytoplasmic acetylation carried out by HAT B (histone acetyltransferase - HAT) – Prepares histones for incorporation into nucleosomes – Acetyl groups later removed in nucleus by deacetylases Histone Acetylation • Nuclear acetylation of core histone N-terminal tails – Catalyzed by HAT A – Attracts bromodomain proteins - essential for transcription – Correlates with transcription activation – Coactivators of HAT A found which may allow loosening of association between nucleosomes and gene’s control region Histone Deacetylation • Transcription repressors bind to DNA sites and interact with corepressors which in turn bind to histone deacetylases – Repressors • Unliganded nuclear receptors • Mad-Max – Corepressors • NCoR/SMRT • SIN3 – Histone deacetylases - HDAC1 and 2 Ternary Protein Complexes • Assembly of complex brings histone deacetylases close to nucleosomes • Deacetylation of core histones allows – Histone basic tails to bind strongly to DNA+ histones in neighboring nucleosomes – This inhibits transcription Activation and Repression Source: Adapted from Wolfe, A.P., 1997. Sinful repression. Nature 387:16-17. Deacetylation of core histones removes binding sites for bromodomain proteins that are essential for transcription activation Chromatin Remodeling • Activation of many eukaryotic genes requires chromatin remodeling • Several protein complexes carry this out – All have ATPase harvesting energy from ATP hydrolysis for use in remodeling – Remodeling complexes are distinguished by ATPase component Remodeling Complexes • SWI/SNF – In mammals, has BRG1 as ATPase – 9-12 BRG1-associated factors (BAFs) • A highly conserved BAF is called BAF 155 or 170 • Has a SANT domain responsible for histone binding • This helps SWI/SNF bind nucleosomes • ISWI – Have a SANT domain – Also have SLIDE domain involved in DNA binding SWI/SNF Chromatin Remodeling Mechanism of Chromatin Remodeling • Mechanism of chromatin remodeling involves: – Mobilization of nucleosomes – Loosening of association between DNA and core histones • Catalyzed remodeling of nucleosomes involves formation of distinct conformations of nucleosomal DNA/core histones when contrasted with: – Uncatalyzed DNA exposure in nucleosomes – Simple nucleosome sliding along a DNA stretch Heterochromatin • Euchromatin: relatively extended and open chromatin that is potentially active • Heterochromatin: very condensed with its DNA inaccessible – Microscopically appears as clumps in higher eukaryotes – Repressive character able to silence genes as much as 3 kb away Heterochromatin and Silencing • Formation of at tips of yeast chromosomes (telomeres) with silencing of the genes is the telomere position effect (TPE) • Depends on binding of proteins – RAP1 to telomeric DNA – Recruitment of proteins in this order: • SIR3 • SIR4 • SIR2 SIR Proteins • Heterochromatin at other locations in chromosome also depends on the SIR proteins • SIR3 and SIR4 interact directly with histones H3 and H4 in nucleosomes – Acetylation of Lys 16 on H4 in nucleosomes prevents interaction with SIR3 – Blocks heterochromatin formation • Histone acetylation also works in this way to promote gene activity Histone Methylation • Methylation of Lys 9 in N-terminal tail of H3 attracts HP1 • This recruits a histone methyltransferase – Methylates Lys 9 on a neighboring nucleosome – Propagates the repressed, heterochromatic state • Methylation of Lys and Arg side chains in core histones can have either repressive or activating effects Modification Interactions • The modifications shown above the tail are activating – Ser phosphorylation – Lys acetylation • Modification below the tail (Lys methylations) is repressive Modification Combinations • Methylations occur in a given nucleosome in combination with other histone modifications: – Acetylations – Phosphorylations – Ubiquitylations • Each particular combination can send a different message to the cell about activation or repression of transcription • One histone modification can also influence other nearby modifications Nucleosomes and Transcription Elongation • An important transcription elongation facilitator is FACT (facilitates chromatin transcription) – Composed of 2 subunits: • Spt16 – Binds to H2A-H2B dimers – Has acid-rich C-terminus essential for these nucleosome remodeling activities • SSRP1 binds to H3-H4 tetramers – Facilitates transcription through a nucleosome by promoting loss of at least one H2A-H2B dimer from the nucleosome • Also acts as a histone chaperone promoting readdition of H2A-H2B dimer to a nucleosome that has lost such a dimer • • • This project is funded by a grant awarded under the President’s Community Based Job Training Grant as implemented by the U.S. Department of Labor’s Employment and Training Administration (CB-15-162-06-60). NCC is an equal opportunity employer and does not discriminate on the following basis: against any individual in the United States, on the basis of race, color, religion, sex, national origin, age disability, political affiliation or belief; and against any beneficiary of programs financially assisted under Title I of the Workforce Investment Act of 1998 (WIA), on the basis of the beneficiary’s citizenship/status as a lawfully admitted immigrant authorized to work in the United States, or his or her participation in any WIA Title I-financially assisted program or activity. Disclaimer • This workforce solution was funded by a grant awarded under the President’s Community-Based Job Training Grants as implemented by the U.S. Department of Labor’s Employment and Training Administration. The solution was created by the grantee and does not necessarily reflect the official position of the U.S. Department of Labor. The Department of Labor makes no guarantees, warranties, or assurances of any kind, express or implied, with respect to such information, including any information on linked sites and including, but not limited to, accuracy of the information or its completeness, timeliness, usefulness, adequacy, continued availability, or ownership. This solution is copyrighted by the institution that created it. Internal use by an organization and/or personal use by an individual for non-commercial purposes is permissible. All other uses require the prior authorization of the copyright owner.