* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Sequence - andreawise
Genetic engineering wikipedia , lookup
Frameshift mutation wikipedia , lookup
Gel electrophoresis of nucleic acids wikipedia , lookup
Genome evolution wikipedia , lookup
DNA vaccination wikipedia , lookup
Zinc finger nuclease wikipedia , lookup
Transposable element wikipedia , lookup
Genealogical DNA test wikipedia , lookup
DNA supercoil wikipedia , lookup
Nucleic acid double helix wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Extrachromosomal DNA wikipedia , lookup
DNA barcoding wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Molecular cloning wikipedia , lookup
Primary transcript wikipedia , lookup
SNP genotyping wikipedia , lookup
United Kingdom National DNA Database wikipedia , lookup
Designer baby wikipedia , lookup
Pathogenomics wikipedia , lookup
Epigenomics wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
DNA sequencing wikipedia , lookup
Human genome wikipedia , lookup
Point mutation wikipedia , lookup
Human Genome Project wikipedia , lookup
Cell-free fetal DNA wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Microevolution wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Whole genome sequencing wikipedia , lookup
Non-coding DNA wikipedia , lookup
History of genetic engineering wikipedia , lookup
Sequence alignment wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Genomic library wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Bisulfite sequencing wikipedia , lookup
Genome editing wikipedia , lookup
Microsatellite wikipedia , lookup
Helitron (biology) wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
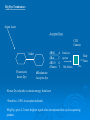
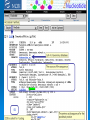
DNA and Beyond 2010 PHS Rashmi Pershad Objectives Learn about the role of DNA sequencing and fragment analysis Learn how to look up information on any gene of interest How to conduct a BLAST search How to go from gene sequence to function The $1,000 Genome DNA Technologies Methodology Fragment Analysis DNA Sequencing Real time PCR Applications: Medical Diagnostics Forensic Analysis Plant Genetics Polymerase Chain Reaction PCR Movie- Cold Spring Harbor http://www.dnalc.org/view/15475-The cycles-of-the-polymerase-chain-reactionPCR-3D-animation-with-no-audio.html PCR Movie Applied Biosystems http://media.invitrogen.com.edgesuite.net/ ab/applications-technologies/pharmabiotherapeutics/pcr.swf Microsatellites What is a microsatellite? Are simple sequence repeats consisting of 1-6 base pair repeats How can they be used? They can be used as genetic markers Study Microsatellite Instability 377 gels 96 well porous comb gels Data from 96 lanes in 2.5 hours Still had to track create size standards and analyze data. Fragment Analysis •Rox 350 size standard 0.02µl per sample •Fiveplex Microsatellite Analysis Limitation of Instrumentation In Cancer Center work with precious archival paraffin embedded patient DNA. Increased sensitivity provides more data from limited sample. Too much sample results in pull up Optimal input of product for us is 0.4 ng/µl DNA Sequencing Advances in DNA Sequencing Part I Overview Maxam Gilbert Sequencing-(1976) Sanger Sequencing Sequencing has been used to map the human genomes, mutation detection study genetic diseases such as CF, MS and to study cancer susceptibility genes. Fluoresent technology In 1986 ABI developed the technology to sequence using 4 dyes in one lane fluorescent technology. This allowed for increase in throughput and removed lane to lane variability. Platforms Initially all the work was performed using slab gels. In the early 1990s 24-36 lane capacity. In 1996 new instruments allowed for higher throughput 48,64 and 96 lane. Now capillary electrophoresis Celera Genomics versus NIH New technology is whole genome sequencing- Helicos Dye Primer Labeling • 4 separate reactions Taq polymerase dNTPs A A ACCG A And ddNTPS Taq polymerase dNTPs C A C ACCG A C And ddNTPS G T ACC G ACCGAC T Dye Terminator labeling A Primer Template AC Taq polymerase, dNTPs + ddTerminators A C G T Advantages •Reaction performed in single tube •Can used unlabeled primers •False stops are undetected ACCG ACCGT Big Dye Terminators Argon Laser Acceptor Dyes CCD Camera dR6G dRox dR110 dTamra linker Fluorescein donor Dye A Emission C spectra G T 500-600nm Raw Data filter dRhodamine Acceptor dye •Donor Dye absorbs excitation energy from laser •Transfers c.100% to acceptor molecule •Big Dye gives 2-3 times brighter signal when incorporated into cycle sequencing product Protocol Sequencing Reagent DNA Primer Big Dye Terminator ready reaction mix 5X reaction Buffer deionised water Total Volume Reaction Protocol Quantity 1.5µl 1.6µl 2µl 3µl 11.9µl 20µl •Set up sequencing reaction in tube. •Keep reagents on ice •Mix well and spin briefly DNA Sequencing Movie DNA Sequencing Movie- Applied Biosystems http://media.invitrogen.com.edgesuite.net/ ab/applications-technologies/pharmabiotherapeutics/DNA_sequencing.swf DATA FROM 3700 GENETIC ANALYZER RUN Raw Sequence Data Start of Sequence Read out to 800 bases Standard Template on Pop4 80cm Capillary. Run under standard conditions BDv3.1 ( 1µl)- 800-900 base reads Data from a 3700 in 4hours run Problematic Template on Pop4 80cm Capillary. GC rich template with and without Enhancer A Big dye Version 3.1 Normal Wilm’s Tumor Exon 9 Mutant C T What do you do once you have a sequence? •Compare it to normal sequence. •Submit it to a sequence alignment program to assemble sequence •Search database to identify sequence DNA Databases When a scientist sequences a segment of DNA, be it a single gene, a gene operon or an entire chromosome or GENOME, the sequence is deposited in an online database. The most commonly used database in this country is NCBI Genbank Other databases include: EMBL-EBI (European Molecular Biology LaboratoryEuropean Bioinformatics Institute) DDBJ (DNA Data Bank of Japan) These databases contains millions of DNA sequences and exchange information daily Bioinformatics Tools BLAST( NCBI TOOL) •Sequence Alignment •Sequence Comparison •Sequence Identification Pub Med PUBMED is the NCBI database of scientific literature that you can search with terms of interest, to see what researchers have discovered about the biology of proteins that are similar to your query sequence. BLAST SEARCH NCBI allows users to search the databases and perform analyses in various ways. You can search by name for nucleotide sequences (genes) or amino acid sequences (proteins). You can search by name for publications about the sequence (recorded in the Science Life literature database called PUBMED). You can search for similar sequences using the feature called BLAST (by inputting all or part of a DNA or amino acid sequence) and compare two or more sequences. NCBI Sequence Record Each sequence record in the NCBI sequence databases is organized into three sections: Header – general information about the sequence including the organism it came from and the paper in which it was first published. Features - information about the role of the sequence in the biology of the organism and any changes that have been made to the sequence. This section also includes information like the length of the sequence, the molecular weight of the protein, and any notes that the depositors wished to add. The start of this section is indicated with the label FEATURES on the left. Terms beginning with a / are referred to as ‘qualifiers’. Examples of qualifiers include /product, /gene, /locus_tag, and /note. Sequence – nucleotides listed in order and numbered. The start of this section is indicated with the label ORIGIN on the left. Reference Material http://www.pseudomonassyringae.org/Outreach/Module_4_Web.htm http://www.digitalworldbiology.com/BLAST /slide1.html Questions? Question 1 1. How can you search Genebank for DNA sequences that are similar to your gene of interest? a) Google b) PubMed c) MySpace d) BLAST Answer D BLAST Question 2 If your E score is = 0 a) The sequence is 100% match b) The sequences are not alike c) There is limited homology between sequences d) Sequences are highly homologous Answer A The e score shows the “expected” match value. The lower the score the more significant the alignment . Question 3 What does PCR stand for? Answer Polymerase Chain Reaction Exercise 1 You will: Copy the sequence provided in the box on the Blast page of the NCBI : websitehttp://www.ncbi.nlm.nih.gov/genome/se q/BlastGen/BlastGen.cgi?taxid=9606 Identify the name of the gene, Provide the title of the publication where the sequence was first published, the name of the journal authors and the full title of the article. What did you learn about this gene? Exercise 1 continued You will be provided with a list of sequences that you can use to perform BLAST searches. For each sequence provided you will answer the questions from the previous slide. If you need to review a BLAST tutorial you can find one at: http://www.digitalworldbiology.com/BLAST/slide1 .html Sequence 1 TCGAAATAACGCGTGTTCTCAACGCGGTCGCGCAGATGCCTTTGC TCATC AGATGCGACCGCAACCACGTCCGCCGCCTTGTTCGCCGTCCC CGTGCCTC AACCACCACCACGGTGTCGTCTTCCCCGAACGCGTCCCGGTC AGCCAGCC TCCACGCGCCGCGCGCGCGGAGTGCCCATTCGGGCCGCAGC TGCGACGGT GCCGCTCAGATTCTGTGTGGCAGGCGCGTGTTGGAGTCTAAA Sequence 2 GTTTATTAGTGATCATGGCTAAGTTTGCGTCCATCATCGCACTT CTTTTT GCTGCTCTTGTTCTTTTTGCTGCTTTCGAAGCACCAACAATGGT GGAAGC ACAGAAGTTGTGCGAAAGGCCAAGTGGGACATGGTCAGGAGT CTGTGGAA ACAATAACGCATGCAAGAATCAGTGCATTAACCTTGAGAAAGC ACGACAT GGATCTTGCAACTATGTCTTCCCAGCTCACAAGTGTATCTGCTA CTTTCC TTGTTAATTTATCGCAAACTCTTTGGTGAATAGTTTTTATGTAAT TTACA CAAAATAAGTCAGTGTCACTATCCATGAGTGATTTTAAGACATG TACCAG ATATGTTATGTTGGTTCGGTTATACAAATAAAGTTTTATTCACCA Sequence 3 CTCGAGACTAGTTCTCTCTCTCTCTCTCTCGTGCCGCATCTCAC ACCTGT GGATGGACGGCAGCTGAACCGCGGGAAACTTTCGTTCTCACTC TACCTAG ATGAACTTTAGTTTATATTAAACACGCGTCGACTCCCACACAAA CCGTGC TCGTTTTACATCTTTGTCTCCGCTTTTGAAAACGAGAAGTTGAA TTCGCA AGACGCAACTTTCCAGCCCCTCACTGAGCGGGCAGAGTCCGT GAAGCGAT GGAGCCGTCCGTCATTCCCGGTGCTGACATACCCGACCTTTAC TCCATTA ACCCGTTTAATGTCACTTTTCCCGACGACGTTTTGAGTTTCGTT CCTGAT GGGAGGAACTACACCGAACCTAACCCGGTAAAGAGCCGCG GAATCATCA TCGCCATTTCCATCACCGCTC Sequence DNA in Thermal Cycler •Place tubes in thermal cycler •Repeat the following for 25 cycles Rapid Thermal Ramp to 96˚C 96˚C for 10 secs Rapid Thermal ramp to50˚C 50˚C for 5 secs Rapid thermal ramp to 60˚C 60˚C for 4minutes •Cool to 4˚C. Hold until ready to purify.