18. The Reference Sequence (RefSeq) Project
... cases that are identified through user input or analysis. The curation process includes analysis of a suite of precomputed BLAST results, literature review, review of available database and Web resources (both in-house and external), and collaboration to curate the nucleotide and protein sequence da ...
... cases that are identified through user input or analysis. The curation process includes analysis of a suite of precomputed BLAST results, literature review, review of available database and Web resources (both in-house and external), and collaboration to curate the nucleotide and protein sequence da ...
Partial report - GEP Community Server
... Note: In some cases, the reconciled gene models (available under "Genes and Gene Prediction Tracks" this "Reconciled Gene GEP UCSC Genome Browser) might incorrect Complete report form for Models" each geneoninthe your project. Copy and paste this form tobe create as because of misannotations or bec ...
... Note: In some cases, the reconciled gene models (available under "Genes and Gene Prediction Tracks" this "Reconciled Gene GEP UCSC Genome Browser) might incorrect Complete report form for Models" each geneoninthe your project. Copy and paste this form tobe create as because of misannotations or bec ...
Human mutations in glucose 6-phosphate dehydrogenase reflect
... order of 70%); when these are compared with those of lower vertebrates, invertebrates, and microorganisms, it is not surprising that the degree of identity decreases but remains higher than 20% (Table 1). The percentage of similarity among all of the 52 sequences ranges between 43 to 98%. A long str ...
... order of 70%); when these are compared with those of lower vertebrates, invertebrates, and microorganisms, it is not surprising that the degree of identity decreases but remains higher than 20% (Table 1). The percentage of similarity among all of the 52 sequences ranges between 43 to 98%. A long str ...
Rfam Documentation
... Seed alignments and secondary structure annotation Our seed alignments are small, curated sets of representative sequences for each family, as opposed to an alignment of all known members. The seed alignment also has as a secondary structure annotation, which represents the conserved secondary struc ...
... Seed alignments and secondary structure annotation Our seed alignments are small, curated sets of representative sequences for each family, as opposed to an alignment of all known members. The seed alignment also has as a secondary structure annotation, which represents the conserved secondary struc ...
Additional file 1
... The same search is performed on Ipmyb1, but no clear consensus can be draw from Jaspar database. The following search is based on the assumption that the binding sites of two collaborative TFs are not far apart (e.g., < 50 bp). 2、 Count appearance number of pattern “CACGTG(…)xxxx” and “xxxx (…)CACGT ...
... The same search is performed on Ipmyb1, but no clear consensus can be draw from Jaspar database. The following search is based on the assumption that the binding sites of two collaborative TFs are not far apart (e.g., < 50 bp). 2、 Count appearance number of pattern “CACGTG(…)xxxx” and “xxxx (…)CACGT ...
Fragaria multicipita - DigitalCommons@University of Nebraska
... standards used were PhiX174 RF HaeIII digest (Life Technologies) and 1.6 kb ladder (Fermentas AB, Vilnius, Lithuania). RFLP patterns were compared with those previously published (Davis et al., 1997; Jomantiene et al. 1998a,b, 2002; Lee et al. 1998). Phytoplasma 16S rRNA group and subgroup designati ...
... standards used were PhiX174 RF HaeIII digest (Life Technologies) and 1.6 kb ladder (Fermentas AB, Vilnius, Lithuania). RFLP patterns were compared with those previously published (Davis et al., 1997; Jomantiene et al. 1998a,b, 2002; Lee et al. 1998). Phytoplasma 16S rRNA group and subgroup designati ...
Computational Definition of
... We have used a computational approach to identify ESEs and ESSs associated with constitutively spliced exons. We focused on constitutive splicing as opposed to alternative splicing so as to tackle the more fundamental problem represented by the former and to avoid what are likely more complex mechan ...
... We have used a computational approach to identify ESEs and ESSs associated with constitutively spliced exons. We focused on constitutive splicing as opposed to alternative splicing so as to tackle the more fundamental problem represented by the former and to avoid what are likely more complex mechan ...
Selecting Degenerate Multiplex PCR Primers
... is to be amplified, two primers, generally referred to as the forward and reverse primers, are needed. In MP-PCR, it is necessary to select a forward and reverse primer for each of the regions to be replicated, and for the large-scale amplification required in SNP Genotyping, there can be hundreds, ...
... is to be amplified, two primers, generally referred to as the forward and reverse primers, are needed. In MP-PCR, it is necessary to select a forward and reverse primer for each of the regions to be replicated, and for the large-scale amplification required in SNP Genotyping, there can be hundreds, ...
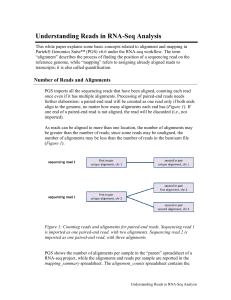
Understanding Reads in RNA-Seq Analysis
... they mapped to two different transcript or different chromosome, they are not compatible with a transcript. Reads that are partially mapped exon reads, intronic reads, intergenic reads are incompatible with any transcript. Incompatible reads do not contribute to gene or transcript level read counts. ...
... they mapped to two different transcript or different chromosome, they are not compatible with a transcript. Reads that are partially mapped exon reads, intronic reads, intergenic reads are incompatible with any transcript. Incompatible reads do not contribute to gene or transcript level read counts. ...
Costimulatory receptors in jawed vertebrates: Conserved
... When relevant genomic regions were identified, potential exons were identified by comparison with known sequences, or from predictions made by Genscan (http://genes.mit.edu/GENSCAN.html). Relevant consensus nucleotide sequences were translated and ORF considered as candidate costimulatory sequences we ...
... When relevant genomic regions were identified, potential exons were identified by comparison with known sequences, or from predictions made by Genscan (http://genes.mit.edu/GENSCAN.html). Relevant consensus nucleotide sequences were translated and ORF considered as candidate costimulatory sequences we ...
APMLE Practice Test II
... Directions for questions 1-56: These questions are followed by four suggested answers. Select the one answer that is best in each case. NOTE: Throughout this test, the term “medial oblique foot” refers to a nonweightbearing medial oblique position in which the film is flat on the orthoposer, the med ...
... Directions for questions 1-56: These questions are followed by four suggested answers. Select the one answer that is best in each case. NOTE: Throughout this test, the term “medial oblique foot” refers to a nonweightbearing medial oblique position in which the film is flat on the orthoposer, the med ...
The Johns Hopkins University - American University of Beirut
... • Did a BLAST search – and you need more info about some of the proteins they found similarities to. • Heard on about a disease gene that was recently discovered, and you want to know more about it. • Want to build a dataset for local blast searches. • A colleague wants you to do an alignment of all ...
... • Did a BLAST search – and you need more info about some of the proteins they found similarities to. • Heard on about a disease gene that was recently discovered, and you want to know more about it. • Want to build a dataset for local blast searches. • A colleague wants you to do an alignment of all ...
Unipro UGENE Manual
... You're going to use only basic UGENE features and don't want to waste Internet traffic You have limited disk space Use the NGS package, if: You're going to analyze ChIP-Seq data with the Cistrome pipeline Explanation of the tip above: Some tools are embedded into UGENE as external. To be launched fr ...
... You're going to use only basic UGENE features and don't want to waste Internet traffic You have limited disk space Use the NGS package, if: You're going to analyze ChIP-Seq data with the Cistrome pipeline Explanation of the tip above: Some tools are embedded into UGENE as external. To be launched fr ...
Unipro UGENE Manual
... You're going to use only basic UGENE features and don't want to waste Internet traffic You have limited disk space Use the NGS package, if: You're going to analyze ChIP-Seq data with the Cistrome pipeline Explanation of the tip above: Some tools are embedded into UGENE as external. To be launched fr ...
... You're going to use only basic UGENE features and don't want to waste Internet traffic You have limited disk space Use the NGS package, if: You're going to analyze ChIP-Seq data with the Cistrome pipeline Explanation of the tip above: Some tools are embedded into UGENE as external. To be launched fr ...
Natural rules for Arabidopsis thaliana
... observed as AG|GUA and UNCAG|GU (N could be any nucleotide) respectively, which is consistent with previous studies [3,12,15]. The results indicated that consensus sequences at splicing sites are important for site recognition but are not enough to define the sites alone. The distributions of A, G, ...
... observed as AG|GUA and UNCAG|GU (N could be any nucleotide) respectively, which is consistent with previous studies [3,12,15]. The results indicated that consensus sequences at splicing sites are important for site recognition but are not enough to define the sites alone. The distributions of A, G, ...
NATIONAL BOARD OF PODIATRIC MEDICAL EXAMINERS
... The Part I Practice Tests are representative of the content covered in the Part I Examination. They include question formats found in the actual examination. They also include questions of varying difficulty. A candidate’s performance on a Practice Test does not guarantee similar performance on the ...
... The Part I Practice Tests are representative of the content covered in the Part I Examination. They include question formats found in the actual examination. They also include questions of varying difficulty. A candidate’s performance on a Practice Test does not guarantee similar performance on the ...
Rapid Diversification of RNase A Superfamily Ribonucleases from
... sequences range in length from 108 to 111 amino acids, and are thus more similar in length to rc-RNase, rc-L1, and rjlec (110 to 111 amino acids) than they are to rc-02, rc-03, rc-04, rc-06, rp-onc, or rp-rap which are all somewhat smaller (104–106 amino acids). The isoelectric points of the five no ...
... sequences range in length from 108 to 111 amino acids, and are thus more similar in length to rc-RNase, rc-L1, and rjlec (110 to 111 amino acids) than they are to rc-02, rc-03, rc-04, rc-06, rp-onc, or rp-rap which are all somewhat smaller (104–106 amino acids). The isoelectric points of the five no ...
METADOMAIN: A PROFILE HMM-BASED PROTEIN DOMAIN
... homology search provides basis for identifying putative genes and assigning those genes to annotated functional categories (e.g. protein domain families). There are two major methods for protein homology search. The first method is based on pairwise sequence alignment. Putative genes can be identified ...
... homology search provides basis for identifying putative genes and assigning those genes to annotated functional categories (e.g. protein domain families). There are two major methods for protein homology search. The first method is based on pairwise sequence alignment. Putative genes can be identified ...
Analysis of Cross Sequence Similarities for Multiple - PolyU
... Most DNA-based compression algorithms rely on encoding together similar repeated regions found within one chromosome sequence. Biocompress proposed by Grumbach and Tahi (1993) is the first algorithm designed specifically for compressing DNA sequences. Both Biocompress and its second version Biocompr ...
... Most DNA-based compression algorithms rely on encoding together similar repeated regions found within one chromosome sequence. Biocompress proposed by Grumbach and Tahi (1993) is the first algorithm designed specifically for compressing DNA sequences. Both Biocompress and its second version Biocompr ...
Title: Statistical Evidence for Common Ancestry
... The Robinson-Foulds distance between trees estimated from nucleotide variation at amino acid invariant sites and from amino acid variation ranged from 2-10 across the 50 data sets (mean =5.6, 39 replicates had RF distance = 4 or 6). Using the exact p-values for significance from table 4 in Hendy et ...
... The Robinson-Foulds distance between trees estimated from nucleotide variation at amino acid invariant sites and from amino acid variation ranged from 2-10 across the 50 data sets (mean =5.6, 39 replicates had RF distance = 4 or 6). Using the exact p-values for significance from table 4 in Hendy et ...
New Allergen Submission Form
... characterized or the new variant is prevalent in a certain region/environment). Use the comments field above. Isoallergens share the following common biochemical properties: similar molecular size, identical biological function, if known, and an amino acid sequence identity above 67%. It is recogniz ...
... characterized or the new variant is prevalent in a certain region/environment). Use the comments field above. Isoallergens share the following common biochemical properties: similar molecular size, identical biological function, if known, and an amino acid sequence identity above 67%. It is recogniz ...
Plastics_and_Reconstructive_Procedures
... other breast, where the same procedure is followed. Once the second side is temporarily closed, the patient is once again sat up to compare both breasts and t determine if further work is needed. If the MD is satisfied, the patient is returned to the supine position and ...
... other breast, where the same procedure is followed. Once the second side is temporarily closed, the patient is once again sat up to compare both breasts and t determine if further work is needed. If the MD is satisfied, the patient is returned to the supine position and ...
pdf
... was filtered onto Sterivex capsules (0.2 µm pore size, Millipore, Inc., Bedford, MA, USA) with a peristaltic pump, quick frozen in liquid nitrogen and stored at −80°C until the DNA could be extracted. DNA analyses are reported for samples from the SNM for the open-ocean locations. The sampling depth ...
... was filtered onto Sterivex capsules (0.2 µm pore size, Millipore, Inc., Bedford, MA, USA) with a peristaltic pump, quick frozen in liquid nitrogen and stored at −80°C until the DNA could be extracted. DNA analyses are reported for samples from the SNM for the open-ocean locations. The sampling depth ...
Minimum SNPs version 2043 user manual
... In MLST the number of nucleotide differences between alleles is ignored and sequences are given different allele numbers whether they differ at a single nucleotide site or at many sites. The rationale is that a single genetic event resulting in a new allele can occur by a point mutation (altering on ...
... In MLST the number of nucleotide differences between alleles is ignored and sequences are given different allele numbers whether they differ at a single nucleotide site or at many sites. The rationale is that a single genetic event resulting in a new allele can occur by a point mutation (altering on ...
Sequence alignment

In bioinformatics, a sequence alignment is a way of arranging the sequences of DNA, RNA, or protein to identify regions of similarity that may be a consequence of functional, structural, or evolutionary relationships between the sequences. Aligned sequences of nucleotide or amino acid residues are typically represented as rows within a matrix. Gaps are inserted between the residues so that identical or similar characters are aligned in successive columns.Sequence alignments are also used for non-biological sequences, such as those present in natural language or in financial data.