* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Document
Mitochondrial DNA wikipedia , lookup
RNA silencing wikipedia , lookup
Point mutation wikipedia , lookup
Extrachromosomal DNA wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Short interspersed nuclear elements (SINEs) wikipedia , lookup
Adeno-associated virus wikipedia , lookup
Microevolution wikipedia , lookup
Genetic engineering wikipedia , lookup
Genome (book) wikipedia , lookup
Messenger RNA wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Pathogenomics wikipedia , lookup
Zinc finger nuclease wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Polyadenylation wikipedia , lookup
Transposable element wikipedia , lookup
History of RNA biology wikipedia , lookup
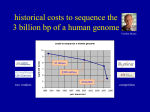
Whole genome sequencing wikipedia , lookup
Non-coding RNA wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Designer baby wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
History of genetic engineering wikipedia , lookup
Epitranscriptome wikipedia , lookup
Helitron (biology) wikipedia , lookup
Non-coding DNA wikipedia , lookup
Human genome wikipedia , lookup
Minimal genome wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Human Genome Project wikipedia , lookup
Genomic library wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Genome evolution wikipedia , lookup
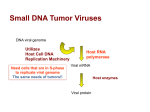
RETROVIRUSES Characteristics • Name originates from the fact that they use reverse transcriptase (retroviruses) • Enveloped virion, 100 nm diameter • Linear +ssRNA genome • 2 identical genomes are packaged in each virion • 7-10 Kb • 7 genera are part of this family including HIV • Diseases they cause: AIDS, leukemia, cancers • A cellular tRNA behaves as primer fro viral genome replication Kaposi’s Sarcoma Kaposis Sarcoma is cancer It is rare More common in AIDS patients Structure Viral Genome • Viral genome exhibits characteristics of cellular mRNA • R sequences-repeated sequences found both at 5’ and 3’ end (~150-200 nt) • U5 region is what keeps the 2 ssRNAs together • PBS-primer binding sequence – In reality it is cellular tRNA that binds this sequence • Downstream the PBS – 3 genes encoding 3 types of proteins – Gag (group specific antigen), pol (polymerase), env (envelope) Viral Entry • Mediated by SU protein • SU interacts with cell surface proteins – In HIV case the CD4 and CCR5 or CXCR4 • Receptor interaction allows for viral entry into cell in 2 ways – Receptor mediated endocytosis followed by virion release via a pH decrease release mechanism – Fusion at plasma membrane, capsid is released into cytosol Viral Entry Conversion of ssRNA Viral Genome Into dsDNA • Reverse transcriptase has 2 distinct activities – 1st is to synthesize DNA – 2nd is to degrade RNA from DNA/RNA molecule • This activity is referred to as ribonuclease H activity • Does not degrade ssRNA Conversion of ssRNA Viral Genome Into dsDNA Conversion of ssRNA Viral Genome Into dsDNA DNA Genome Is Integrated Into Cellular Genome • • • • Insertion sites are random Enzyme responsible for insertion is Integrase Integrase is found in the core of the virion Integrase binds the 2 ends of the viral dsDNA genome and brings them together DNA Genome Is Integrated Into Cellular Genome • • • • Enzyme targets phosphodiester bonds for cleavage/insertion 2 hanging nucleotides are removed 4-6 nt of host ssDNA is matched and ligation site is fixed entirely Loss of 2 nt from viral DNA is insignificant Proviral DNA Will Be Expressed At Any Time In The Future • • • • • • • • • The appropriate transcription factors are needed for expression of inserted genome to begin U3 region is the binding site for a number of cellular transcription factors A TATA box is present upstream (U3/R segments) allowing transcription initiation to begin by RNA Pol II Transcription begins at the junction of U3/R and proceeds through the whole genome A Poly(A) signal directs cleavage of transcript at R/U5 junction RNA is polyadenylated by cellular enzymes RNA transcript generated is identical to initial infecting RNA genome Despite the fact that 2 LTR exist at the ends of proviral DNA, transcription begins only at left side It is thought to be due to Promoter occlusion – RNA Pol II displaces transcription factors on the right • In similar way polyadenylation only occurs to the right – AAUAAA (poly A signal sequence) is also present on the left Differential Splicing Generates Multiple mRNAs • Transcription produces genome length mRNA • The different viral proteins are produced from this mRNA after it is spliced • At least 2 types of mRNAs are produced in retroviruses – 1 unspliced (whole genome) • Used for gag and gag/pol proteins – 1 spliced (gag and gag/pol is removed) • Only env proteins are produced from this mRNA • Some retroviruses have more elaborate splicing schemes – Ex. Lentiviruses (HIV), Rous Sarcoma virus Differential Splicing Generates Multiple mRNAs • HIV overcomes stop codon resulting in translation of gag/pol protein • It achieves that by shifting ribosome at a precise position prior to termination codon • This way it avoids stop codon and addresses the fact that pol protein has a different reading frame • HIV and some other retroviruses achieve this SHIFTING by making use of heptamers such as UUUUUUA (HIV-1) • Why such a scheme? – To ensure right ratio of gag to gag/pol proteins for generating virions Retrovirus Based Gene Therapy • • • The ability of retroviruses to permanently introduce genes into host genome make them good candidates for gene therapy to fix mutated genes or introduce new genes One major issue is to ensure that no tumors are created Engineered retroviruses are missing gag/pol/env genes – This ensures no viral replication – Provides space for new gene • Engineered virus is prepared by packaging recombinant RNA into packaging cell lines (they are missing RNA packaging signals) – Vector is expressed in packaging cell line and gets packaged into virions • Target cells are mixed with virus – A selection gene (neomycin resistant gene) is used to eliminate cells that are not infected • • • In the past the limitation was that they could only infect dividing cells Now lentiviruses (HIV) are used which can infect non-dividing differentiated cells One limitation is the size of the gene – Up to 10 Kb gene size can be inserted in the genome