* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Tutorial 7 – Secretory Pathway
Chloroplast DNA wikipedia , lookup
Protein (nutrient) wikipedia , lookup
Protein phosphorylation wikipedia , lookup
Protein moonlighting wikipedia , lookup
Model lipid bilayer wikipedia , lookup
Cytokinesis wikipedia , lookup
Magnesium transporter wikipedia , lookup
Protein structure prediction wikipedia , lookup
Intrinsically disordered proteins wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Cell nucleus wikipedia , lookup
SNARE (protein) wikipedia , lookup
Cell membrane wikipedia , lookup
Western blot wikipedia , lookup
Signal transduction wikipedia , lookup
Proteolysis wikipedia , lookup
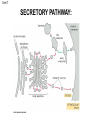
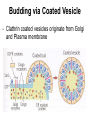
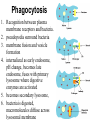
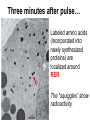
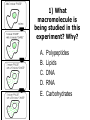
Unit 7 Endomembranes Unit 7 SECRETORY PATHWAY: Secretory Pathway • Proteins are synthesized on the Rough ER. • Move via vesicles to Golgi • Move via vesicles to Plasma membrane - Budding is through coated vesicles (ie. clathrin) - Docking is through the SNAREs Budding via Coated Vesicle - Clathrin coated vesicles originate from Golgi and Plasma membrane Docking via SNARES Vesicles are targeted by interactions between vSNARES and t-SNARES • each vesicle has unique v-SNARES embedded in its membrane • each target membrane has unique t-SNARES (vSNARE receptors) embedded in the target membranes Protein Transport: 3 mechansims 1. Nuclear import 2. Protein translocators - protein pores in the membrane that help unfolded proteins get into membranes 3. Transport vesicles - membrane vesicles pinch off a donor compartment, fuse with a recipient compartment and carry both: - soluble proteins in lumen and - membrane proteins associated with bilayer Protein Targeting • Sorting signals are necessary to direct a protein to a particular organelle. 1. 2. 3. 4. Nuclear localization signal ER signal sequence KDEL - retention in the ER lumen Mitochondria/chloroplast signal sequence NOTE: Proteins destined for nucleus, chloroplast, mitochondria, and peroxisome are made in cytosol on free ribosomes! Nuclear localization signal - On soluble, folded nuclear proteins, made on free ribosomes in the cytosol - Contains one or two short sequences with positively charged amino acids - Bind Nuclear import receptors in cytoplasm - Move into nucleus through nuclear pores ER Signal Sequence - Protein Translocator = at N terminal - Consists of hydrophobic amino acids. - Causes proteins meant for secretory pathway to enter the ER - Retention signal = KDEL - Used to target “resident proteins” of ER to get back to ER from Golgi after sorting Mitochondria/chloroplast Signal Sequence - - Protein translocator for proteins made on free ribosomes destined for mitochondria and chloroplast signal sequence at N terminal What if the sequence is moved to C terminal? Unit 7 Models of Golgi Function There are two competing theories: Cisternal progression model: New cisternae form continuously from ER vesicles. Cisternae move through the stack from cis to trans and finally break up into transport vesicles at the trans face. Vesicle transport model: Cisternae remain fixed. Both membrane and content move from the cis to the trans cisternae in transport vesicles. Unit 7 Endocytosis - protein binds receptors on the cell surface and is internalized in clathrin-coated vesicles. - the vesicles lose their coats and fuse with endosomes. - Cargo protein separates from its receptor in endosome. - protein is transferred to a lysosome and degraded to release free cholesterol - protein receptors return to the plasma membrane via transport vesicles Phagocytosis 1. Recognition between plasma membrane receptors and bacteria. 2. pseudopodia surround bacteria 3. membrane fusion and vesicle formation 4. internalized as early endosome, pH change, becomes late endosome, fuses with primary lysosome where digestive enzymes are activated 5. becomes secondary lysosome, 6. bacteria is digested, macromolecules diffuse across lysosomal membrane Pulse-Chase Autoradiography An experimental approach to observing protein secretion What is Autoradiography? • Autoradiography is the use of radioactively labeled molecules to look at cell processes. • A labeled molecule can be located because its radioactivity develops the silver grains on a photographic emulsion. “Tailing” a labeled molecule • Allows us to observe the movement of molecules through the cell over time. • Cellular pathways are revealed as the progress of molecules is monitored. The “Pulse” and the “Chase” • The "Pulse" consists of radioactive material added for a very brief period and then washed away. • Then the “Chase” begins- non-radioactive molecules are added. • This creates a group of labeled molecules, with unlabeled molecules in front and behind, that move through a particular metabolic pathway. Now for a demonstration! • Stomach cells which secrete digestive enzymes (zymogen) are supplied with radioactively labeled amino acids. • After a brief period -the “Pulse” - the excess labeled amino acids are washed away. • The cells are then supplied with unlabeled amino acids -the “Chase” And in the TEM we see… Three minutes after pulse… Labeled amino acids (incorporated into newly synthesized proteins) are localized around RER. The “squiggles” show radioactivity. Seven minutes after pulse… • The majority of the newly synthesized proteins have moved to the periphery of the Golgi complex. • Arrows indicate periphery of Golgi complex. 37 minutes after pulse… • Labeled proteins are concentrated over secretory vesicles called condensing vacuoles (CV). • Arrows indicate periphery of Golgi complexes. • Secretory vesicles containing zymogen are marked Z. 117 minutes after pulse… • Radioactivity is mainly localized over secretory vesicles containing zymogen. • Some labeled protein has already been secreted into the lumen (L) . Where are they now? (CV) (Z) • Samples are taken after various time periods and the location of the labeled molecules is identified. Got it? Feed the cell a pulse of radioactively labeled molecules and see what it does with them. So here are a few questions… 1 2 3 4 1. PULSE: Cell is exposed to radioactively-labelled nucleotides (green) for 3 minutes. The nucleotides are taken up by the nucleus. 1 2 3 4 2. CHASE: The cell is exposed to an excess of non-radioactive nucleotides. After five minutes of chase, some radioactive molecules are found in the cytoplasm. 1 2 3 4 3. 15 MINUTES: Most of the radioactivity has moved from the nucleus to the cytoplasm after 15 minutes of the chase. 1 2 3 4 4. 30 MINUTES: After 30 minutes, all of the radioactive label is found outside the nucleus. 1) What macromolecule is being studied in this experiment? Why? A. B. C. D. E. Polypeptides Lipids DNA RNA Carbohydrates 2. Where is this macromolecule being synthesized? A. B. C. D. E. Nucleus Golgi Rough ER Cytosol Smooth ER 3. After thirty minutes, is the macromolecule still being synthesized? 4) Which is the best explanation for the observations? A. Synthesis doesn't continue as there is no more labeled nucleotide. B. Synthesis doesn't continue as the cell is not making any more RNA. C. You cannot 'see' that synthesis is continuing as the nucleotides being incorporated are not labeled and hence can not be visualized by autoradiography. NEXT TUTORIAL Structure and function of the CYTOSKELETON – Actin Microfilaments – Microtubules – Intermediate Filaments