* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download notes File - selu moodle
Epigenetics of neurodegenerative diseases wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Epigenomics wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Transcription factor wikipedia , lookup
Human genome wikipedia , lookup
Oncogenomics wikipedia , lookup
DNA polymerase wikipedia , lookup
Genome evolution wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Designer baby wikipedia , lookup
Cell-free fetal DNA wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Short interspersed nuclear elements (SINEs) wikipedia , lookup
RNA interference wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Epigenetics of human development wikipedia , lookup
History of genetic engineering wikipedia , lookup
Non-coding DNA wikipedia , lookup
RNA silencing wikipedia , lookup
Nucleic acid tertiary structure wikipedia , lookup
Microevolution wikipedia , lookup
Helitron (biology) wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Polyadenylation wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Deoxyribozyme wikipedia , lookup
History of RNA biology wikipedia , lookup
Frameshift mutation wikipedia , lookup
Expanded genetic code wikipedia , lookup
Messenger RNA wikipedia , lookup
Point mutation wikipedia , lookup
Non-coding RNA wikipedia , lookup
Transfer RNA wikipedia , lookup
Genetic code wikipedia , lookup
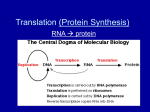
Chapter 15 Vocabulary: One gene / one polypeptide hypothesis Central Dogma Transcription Translation mRNA tRNA rRNA snRNA SRP microRNA Codon Intron Exon Point mutation Frameshift mutation Translocation Outline 15.1 Nature of Genes Garrod examined diseases that seemed to run in families. He hypothesized that a mutation in a gene affected an enzyme (a gene product) and that these mutations could be passes on. Beadle and Tatum induced DNA damage that altered the functionality of their enzyme product. They were then able to verify that these mutations could be passed on in a Mendelian fashion. Since they observed that single gene mutations affected single enzymes involved in a metabolic pathway this lead to the one gene / one polypeptide hypothesis. The central dogma of molecular biology explains how the genotype gives rise to the phenotype of an organism. Simplistically DNA RNA proteins Transcription translation Modified with discovery of reverse transcriptase (found in retroviruses) DNA ↔ RNA proteins Transcription uses the template strand of DNA to make a mRNA strand that has the same sequence as the coding strand (except that there are U’s in RNA and T’s in DNA) Translation uses a ribosome to read the mRNA and synthesize proteins RNA’s mRNA – carries the code rRNA – component of ribosomes tRNA – carries amino acids used to build proteins snRNA – involved in processing mRNA transcript SRP – allows for translation on the roughER MicroRNA – regulatory function siRNA – regulatory function 15.2 Genetic Code Crick and Brenner discovered that a triplet of nucleotides, codon, encodes for a single amino acid by inducing mutations that deleted single, double and triple nucleotides from DNA and observing the protein products. 43 Genetic code is degenerate (redundant) Stop codons: UAG, UAA, UGA Start codon: AUG (methionine) Wobble effect at third position Near universal (exceptions in organelles and ciliates (prokaryotes also use Nformylmethionine instead of methionine to initiate translation) 15.3 Prokaryotic Transcription Promoter – sequence within DNA Docking site for RNA polymerase Signifies start of a gene Infers directionality of the gene Elongation uses RNA polymerase to add ribonucleotides that are complementary to the template strand of DNA RNA polymerase has a subunit that recognizes the promoter with accuracy and another subunit that synthesizes mRNA. RNA polymerase does not require a primer Most common mechanism for termination is the formation of a hairpin structure In prokaryotes transcription and translation happen simultaneously. Operons are multiple genes under the control of the same promoter 15.4 Eukaryotic Transcription Promoter - -10sequence is a TATA box More initiation factors involved Elongation occurs in the same fashion, but eukaryotes have multiple RNA polymerases RNA polymerase I – transcribes rRNA RNA polymerase II – transcribes mRNA and snRNA RNA polymerase III – transcribes tRNA and other small RNA Termination sites are not well defined mRNA processing: 5’ 7-methyl guanosine cap is added Allows for binding to the ribosome Protects end from degradation 3’ poly A tail is added Protects end from degradation Regulates how many products can be made from a single transcript Intron splicing 15.5 Eukaryotic pre-mRNA splicing Introns Exons (1-1.5% of the genome) snRNA recognizes intron exon junction and form a splicesome (introns are tagged by their sequence) Cleavage occurs at 5’ end of intron and a lariat is formed Free 3’ end of exon is used to displace the intron and join exon to exon Alternate splicing allows a single transcript to be translated into different proteins. (Very common in the immune system – VDJ recombination allows for the creation of endless variety in antibodies.) 15.6 Structures of tRNA and Ribosomes tRNA has two ends anticodon loop acceptor end to which an amino acid is bound by aminoacyl-tRNA synthetases Once tRNA has bound amino acid it is “charged” Ribosomes are composed of large and small subunits Small subunit binds mRNA Large subunit has enzyme activity that will bond amino acids Ribosome has 3 sites: A – accepts new charged tRNAs with anticodons that are complementary to the mRNA P – site where the formation of a peptide bond between amino acids is catalyzed E – site where empty tRNA is released from ribosome 15.7 Process of Translation Prokaryotic initiation Small subunit binds mRNA at conserved sequence Initiation factors associate Initiator tRNA enters ribosome at start codon N-formymethionine is first amino acid added. Eukaryotic initiation Small subunit binds to 5’ cap Methionine is first amino acid added Elongation tRNA with anticodon that can bind codon enters A site (wobble effect) peptide bond formed in P site empty tRNA leaves ribosome translocation repeat Termination Stop codon is reached Release factors enter ribosome instead of tRNA Direction to roughER for translation involved signal sequence in beginning of polypeptide. SRP bind polypeptide and roughER receptor and translation occurs into roughER. 15.8 Summarizing Gene Expression 15.9 Mutations: Altered Genes Mutation – any change in the DNA sequence of an organism Point mutation – insertion, deletion, substitution Substitution Silent Missense Transitions Transversions Nonsense Insertion and deletion Frameshift Triplet repeat expansion mutations Chromosomal mutations Deletions – portion lost Duplication – portion is copied Inversions – portion excised, inverted, inserted Translocation – portion moved from one locus to another Problems with regulation Problems with meiosis This is really about 3 chapters in one chapter; transcription, translation and them mutations. Similar to DNA replication, the overall process is very important for them to understand. Any specific questions will be about eukaryotes rather than prokaryotes, but pointing out differences is useful.