* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download problem set
Deoxyribozyme wikipedia , lookup
Genome evolution wikipedia , lookup
Neuronal ceroid lipofuscinosis wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Protein moonlighting wikipedia , lookup
Gene desert wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Gene therapy wikipedia , lookup
Transfer RNA wikipedia , lookup
Epigenetics in learning and memory wikipedia , lookup
Microevolution wikipedia , lookup
Adeno-associated virus wikipedia , lookup
Gene nomenclature wikipedia , lookup
Nucleic acid tertiary structure wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Short interspersed nuclear elements (SINEs) wikipedia , lookup
Point mutation wikipedia , lookup
Designer baby wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Helitron (biology) wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Gene expression profiling wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
History of RNA biology wikipedia , lookup
RNA silencing wikipedia , lookup
Polyadenylation wikipedia , lookup
Mir-92 microRNA precursor family wikipedia , lookup
RNA interference wikipedia , lookup
Non-coding RNA wikipedia , lookup
RNA-binding protein wikipedia , lookup
Messenger RNA wikipedia , lookup
Alternative splicing wikipedia , lookup
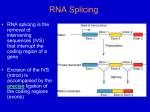
Chap. 8 Problem 1 Mechanisms of post-transcriptional gene control of protein coding genes are shown in Fig. 8.1. The most commonly used mechanism is the regulation of alternative pre-mRNA splicing. However, other methods such as regulation of mRNA decay and translation inhibition by miRNA can play important regulatory roles, depending on the gene. Chap. 8 Problem 3 The sequences in a pre-mRNA that dictate where splicing occurs are located at the exon/intron boundaries of the message (Fig. 8.7 below). These sequences are bound by the snRNA components of the snRNPs that make up spliceosomes (Fig. 8.9). Thus, the intron sequences ultimately tell the splicing machinery where to process the mRNA. The branch point A residue of the intron carries out the first transesterification reaction in which the phosphodiester bond between the upstream exon and the intron is broken (Fig. 8.8). The 2’ OH group of the A residue is free to carry out this reaction. The 3’ OH group of the A residue is linked in a phosphodiester bond. Chap. 8 Problem 4 Heterogeneous nuclear RNAs (hnRNA) include several different types of RNA found in the nucleus, including pre-mRNAs and processing intermediates. Small nuclear RNAs (snRNA) participate in splicing reactions. The structures and functions of the U1 and U2 snRNAs are shown in Fig. 8.9. Micro RNAs (miRNAs) and short interfering RNAs (siRNAs) are involved in posttranscriptional gene silencing. miRNAs inhibit translation of mRNA, while siRNAs cause mRNA degradation (Fig. 8.25). Chap. 8 Problem 5 Both group II intron self-splicing and spliceosome-mediated splicing occur by similar transesterification reactions and produce similar products (Fig. 8.14). In group II intron splicing all of the reactions are carried out by the folded intron itself; in spliceosome-mediated splicing, protein and snRNA components assemble to create the functional enzymatic structure. Evidence supporting the idea that introns in pre-mRNAs evolved from group II self-splicing introns comes from experiments showing that group II self-splicing can be reconstituted using a combination of separate intron fragments, and does not require an intact intron per se. Chap. 8 Problem 6 The results suggest that the fifth intron is not efficiently spliced out of the pre-mRNA transcript for this gene in most tissues. This results in truncation of the transcript at the cleavage and polyadenylation site in the fifth intron. However, the splicing of the fifth intron is efficient in muscle cells. This removes the cleavage and polyadenylation signal and results in a longer spliced mRNA containing all 10 exons. A muscle-specific splicing factor may be responsible for removal of the fifth intron in these cells. This factor may participate in the assembly of cross-exon recognition complexes bound to the fifth and sixth exons (Fig. 8.13). Chap. 8 Problem 10 The function of a gene can be knocked down (inhibited) by the technique of RNA interference (RNAi). In this method a doublestranded siRNA of 21-23 nucleotides, in which the two bases at each 3’ end are single-stranded, is introduced into a cell. There, it forms a complex with the RISC (Fig. 8.26) which base-pairs it to its target mRNA. Because base-pairing is exact, the mRNA is cleaved by the Argonaute protein of the RISC (Fig. 8.25). The fragments of the cleavage event then are rapidly degraded by the cell, and the mRNA is not translated.