* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download PCR and diagnostics II
Human genome wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Zinc finger nuclease wikipedia , lookup
Epigenomics wikipedia , lookup
Metagenomics wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
BRCA mutation wikipedia , lookup
Primary transcript wikipedia , lookup
Genome (book) wikipedia , lookup
Transposable element wikipedia , lookup
Genomic library wikipedia , lookup
Genetic engineering wikipedia , lookup
Cancer epigenetics wikipedia , lookup
Molecular Inversion Probe wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Genome evolution wikipedia , lookup
Genetic code wikipedia , lookup
Population genetics wikipedia , lookup
History of genetic engineering wikipedia , lookup
Koinophilia wikipedia , lookup
Bisulfite sequencing wikipedia , lookup
Neuronal ceroid lipofuscinosis wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Designer baby wikipedia , lookup
Non-coding DNA wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Saethre–Chotzen syndrome wikipedia , lookup
Deoxyribozyme wikipedia , lookup
SNP genotyping wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Cell-free fetal DNA wikipedia , lookup
Genome editing wikipedia , lookup
Oncogenomics wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Helitron (biology) wikipedia , lookup
Microevolution wikipedia , lookup
Microsatellite wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
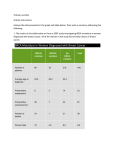
TYPES OF MUTATION CAUSING HUMAN GENETIC DISEASE Nucleotide substitutions (point mutations) Missense mutations Nonsense mutations Spice site mutations Frame shift mutations Small, a few base pairs Rearrangements Deletions Insertions Duplications Unstable repeat sequences Large, can be many kilobases MUTATION Mutations in Coding regions Mutations in regulatory domains Protein abnormal Expression abnormal Loss of function - common Gain of function - rare GENETIC TESTING IN HEMOPHILIA A X-linked recessive, 1/10000 males variable in severity severe cases, spontaneous and life threatening bleeding repeated episodes can cause joint deformity and crippling treatable Gene is large, many mutations, can look for specific common inversion with a PCR based test. • e.g. Alpha anti trypsin deficiency •Disease leads to increased probability of developing pulmonary emphysema •Results from single base pair change at a known nucleotide position • Synthetic oligonucleotide probe that contains the wild type sequence in the relevant region of the gene can be used in a Southern blot analysis to determine whether the DNA contains wild type or mutant sequence • Using the appropriate temperature the complimentary sequence will hybridize but a sequence with even a single mismatched base will not. E.g. Oligoligation assay • Normal sequence pair at site AT, Mutant is GC • 2 short oligonucleotides that are complimentary to one of the 2 native DNA strands are synthesised • Probe X has as its last base at the 3’ end the nucleotide that is complimentary to the normal sequence. It does not hybridize well to the mutant sequence as there is a mismatch • Run test sample next to normal control • Oligos hybridize • When ligase is added the oligos bound to the mutant can’t ligate as have wobbley base that is misaligned • In order to see whether the single base mutation is present need to be able to distinguish between ligated and non ligated (containing mutation) • Probe X has a biotin residue or fluorescent molecule at the 5’ end, Probe Y has a dioxygenin residue at the 3’ end (called PEO in diagram) • After hybridization and ligation, DNA is denatured to release hybridization probes and mix is transferred to wells coated with streptavidin. Biotin binds • Unbound material washed away • Anti dioxygenin antibodies coupled to alkaline phosphatase added to the well • Substrate for AP added • If oligos ligated get colour in well • If oligos not ligated no colour • Some genetic diseases can be attributed to multiple mutations (together or separately) at multiple different sites. PCR has played a tremendous role in making diagnosis possible in a timely fashion. E.g. BRCA 1 •Plays a role in hereditary breast cancer • Very long gene • Analysis is PCR based • Don’t know specifically what you are looking for e.g. can be any of many mutations in BRCA 1, not all yet identified • BRCA 1 has 24 exons that span a huge number of bases • Most mutations have been found in Exon 11 BRCA 1 Exon 11 • 3500bases long • Too long for convenient PCR, split into 3 pieces for analysis • Oligonucleotides incorporate a promoter so amplified products end up with a promoter on the front • 3 pieces are amplified and each used separately in a transcription translation system • Protein products are produced • Run on PAGE to see if are the correct size (next to normal controls) • If incorrect size know there was a mutation • Screening of the protein product allows screening of a very large pieceof DNA when you don’t know specifically what you are looking for Exons 2-10 • ReverseTranscriptase PCR is used • 2-10 includes many introns and far too long to amplify. If start with RNA there are nointrons • Use RT to get DNA, amplify with special oligo with promoter sequences • Use transcription translation system as before, look at protein product size Exons 12-24 • As exons 2-12 Exon 2 • Mutation found that is common in Ashkenazi Jews • If know source sometimes just screen directly from genomic DNA, and look at size on gel to see if different from normal control. Fragile X Syndrome 1/1200 males, 1/2500 females - most common form of X-linked mental retardation Varying degrees of developmental delay Hypermobile joints High arched palate Long face Large protruding ears •Known to be caused by expansion of a CGG repeat in the 5’ end of the FMR-1 gene. •The repeat is polymorphic in the general population 6-45 CGG repeats •Carriers, male and female, unaffected carry 50 to 200 CGGrepeats •Affected individuals carry greater than 200 CGG repeats •When the repeat size is great than 200, expression of the FMR-1 gene is turned off •The presence of more than 200 CGG repeats in the FMR-1 gene contributes to the •fragility of the X chromosome and this can be observed cytogenetically. FMR-1 CGG Repeat Southern blot Affected >200 repeats Premutation 60-200 repeats EcoRI BssHII * EcoRI CGGn