* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Inquiry into Life Twelfth Edition
Gel electrophoresis of nucleic acids wikipedia , lookup
Gene expression programming wikipedia , lookup
Genealogical DNA test wikipedia , lookup
DNA damage theory of aging wikipedia , lookup
Zinc finger nuclease wikipedia , lookup
Minimal genome wikipedia , lookup
DNA polymerase wikipedia , lookup
Nucleic acid tertiary structure wikipedia , lookup
Cancer epigenetics wikipedia , lookup
Copy-number variation wikipedia , lookup
Metagenomics wikipedia , lookup
Genetic engineering wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Nucleic acid double helix wikipedia , lookup
Cell-free fetal DNA wikipedia , lookup
Epigenomics wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Human genome wikipedia , lookup
DNA supercoil wikipedia , lookup
RNA silencing wikipedia , lookup
Short interspersed nuclear elements (SINEs) wikipedia , lookup
Molecular cloning wikipedia , lookup
DNA vaccination wikipedia , lookup
Genomic library wikipedia , lookup
Non-coding RNA wikipedia , lookup
Genome evolution wikipedia , lookup
History of RNA biology wikipedia , lookup
Designer baby wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Extrachromosomal DNA wikipedia , lookup
Point mutation wikipedia , lookup
Genome editing wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Microevolution wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
History of genetic engineering wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Non-coding DNA wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Primary transcript wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
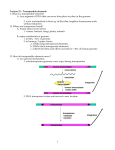
Lecture PowerPoint to accompany Molecular Biology Fourth Edition Robert F. Weaver Chapter 23 Transposition Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. 23.1 Bacterial Transposons • A transposable element moves from one DNA address to another • Originally discovered in maize, transposons have been found in all kinds of organisms – Bacteria – Plants – Humans 23-2 Discovery of Bacterial Transposons • • • • • Phage coat is made of protein Always has the same volume DNA is much denser than protein More DNA in phage, denser phage Extra DNAs that can inactivate a gene by inserting into it were the first transposons discovered in bacteria • These transposons are called insertion sequences (ISs) 23-3 Insertion Sequences • Insertion sequences are the simplest type of bacterial transposon • They contain only the elements necessary for their own transposition – Short inverted repeats at their ends – At least 2 genes coding for an enzyme, transposase that carries out transposition • Transposition involves: – Duplication of a short sequence in the target DNA – One copy of this sequence flanks the insertion sequence on each side after transposition 23-4 Generating Host DNA Direct Repeats 23-5 Complex Transposons • The term “selfish DNA” implies that insertion sequences and other transposons replicate at the expense of their hosts, providing no value in return • Some transposons do carry genes that are valuable to their hosts, antibiotic resistance is among most familiar 23-6 Antibiotic Resistance and Transposons • Donor plasmid has Kanr, harboring transposon Tn3 with Ampr • Target plasmid has Tetr • After transposition, Tn3 has replicated and there is a copy in target plasmid • Target plasmid now confers both Ampr, Tetr 23-7 Transposition Mechanisms • Transposons are sometimes called “jumping genes”, DNA doesn’t always leave one place for another • When it does, nonreplicative transposition – “Cut and paste” – Both strands of original DNA move together from 1 place to another without replicating • Transposition frequently involves DNA replication – – – – 1 copy remains at original site New copy inserts at the new site Replicative transposition “Copy and paste” 23-8 Replicative Transposition of Tn3 • In first step, 2 plasmids fuse, phage replication, forms a cointegrate – coupled through pair of Tn3 copies • Next is resolution of cointegrate, breaks down into 2 independent plasmids, catalyzed by resolvase gene product 23-9 Detailed Tn3 Transposition 23-10 Nonreplicative Transposition • Starts with same 2 first steps as in replicative transposition • New nicks occur at arrow marks • Nicks liberate donor plasmid minus the transposon • Filling gaps and sealing nicks completes target plasmid and its new transposon 23-11 23.2 Eukaryotic Transposons • Transposons have powerful selective forces on their side • Transposons carry genes that are an advantage to their hosts – Their host can multiply at the expense of completing organisms – Can multiply the transposons along with rest of their DNA • If transposons do not have host advantage, can replicate themselves within their hosts 23-12 Examples of Transposable Elements • Variegation in the color of maize kernels is caused by multiple reversions of an unstable mutation in the C locus, responsible for kernel color • Mutation and its reversion result from Ds (dissociation) element – Transposes into the C gene – Mutates it – Transposes out again, revert to wild type 23-13 Ds and Ac of Maize • Ds cannot transpose on its own • Must have help from an autonomous transposon, Ac (for activator) – Ac supplies transposase – Ds is an Ac element with most of its middle removed – Ds needs • A pair of inverted terminal repeats • Adjacent short sequences that Ac transposase can recognize 23-14 Transposable Elements in Maize 23-15 Structures of Ac and Ds 23-16 P Elements • The P-M system of hybrid dysgenesis in Drosophila is caused by conjunction of 2 factors: – Transposable element (P) contributed by the male – M cytoplasm contributed by the female allows transposition of the P element • Hybrid offspring of P males and M females suffer multiple transpositions of P element • Damaging chromosomal mutations are caused that render the hybrids sterile • P elements have practical value as mutagenic and transforming agents in genetic experiments with Drosophila 23-17 23.3 Rearrangement of Immunoglobulin Genes • Mammalian genes use a process that closely resembles transposition for: – B cell antibodies – T cell receptors • Recombinases involved in these processes have similar structures 23-18 Antibody Structure • Antibody is composed of 4 polypeptides – 2 heavy chains – 2 light chains • Sites called variable regions – Vary from 1 antibody to another – Gives proteins their specificity • Rest of protein is constant region 23-19 Immune System Diversity • Enormous diversity of immune system is generated by 3 basic mechanisms: – Assembling genes for antibody light chains and heavy chains from 2 or 3 component parts – Joining the gene parts by an imprecise mechanism that can delete bases or add extra bases – Causing a high rate of somatic mutations, probably during proliferation of a clone if immune cells 23-20 Rearrangement of Antibody Light Chain Gene 23-21 Antibody Heavy Chain Coding Regions Human heavy chain is encoded in – – – – 48 variable segments 23 diversity segments 6 joining segments 1 constant segment 23-22 Recombination Signals • The recombination signal sequences (RSSs) in V(D)J recombination consist of: – Heptamer – Nonamer – Separated by 12-bp or 23-bp spacers • Recombination occurs only between a 12 signal and a 23 signal • Guarantees that only 1 of each coding region is incorporated into the rearranged gene 23-23 The Recombinase • Recombination-activating gene (RAG-1) stimulated V(D)J joining activity in vivo • Another gene tightly liked to RAG-1 also works in V(D)J joining, RAG-2 • These genes, RAG-1 and RAG-2, are expressed only in pre-B and pre-T cells 23-24 Mechanism of V(D)J Recombination • RAG1 and RAG2 introduce single-strand nicks into DNA adjacent to either a 12 signal or 23 signal • Results in transesterification where newly created 3’-OH group: – Attacks the opposite strand – Breaks it – Forms hairpin at the end of the coding segment • Hairpins then break in an imprecise way that allows joining of coding regions with loss of bases or gain of extra bases 23-25 23.4 Retrotransposons • Retrotransposons replicate through an RNA intermediate • Retrotransposons resemble retroviruses – Retroviruses can cause tumors in vertebrates – Some retroviruses cause diseases such as AIDS • Before studying retrotransposons, look at replication of the retroviruses 23-26 Retroviruses • Class of virus is named for its ability to make a DNA copy of its RNA genome • This reaction is the reverse of the transcription reaction – reverse transcription • Virus particles contain an enzyme that catalyzes reverse transcription reaction 23-27 Retrovirus Replication • Viral genome is RNA, with long terminal repeats at each end • Reverse transcriptase makes linear, ds-DNA copy of RNA • ds-DNA copy integrates back into host DNA = provirus • Host RNA polymerase II transcribes the provirus to genomic RNA • Viral RNA packaged into a virus particle 23-28 Model for Synthesis of Provirus DNA • RNase H degrades the RNA parts of RNA-DNA hybrids created during the replication process • Host tRNA serves as primer for reverse transcriptase • Finished ds-DNA copy of viral RNA is then inserted into the host genome • It can be transcribed by host polymerase II 23-29 Retrotransposons • Several eukaryotic transposons transpose in a way similar to retroviruses – Ty of yeast – copia of Drosophila • Start with DNA in the host genome – Make an RNA copy – Reverse transcribe it within a virus-like particle into DNA that can insert into new location • HERVs likely transposed in the same way until ability to transpose lost – HERV = human endogenous retroviruses 23-30 Ty Transcription 23-31 Non-LTR Retrotransposons • LTR are lacking in most retrotransposons • Most abundant type lacking LTR are LINEs and LINE-like elements – Long interspersed elements – Encode an endonuclease that nicks target DNA – Takes advantage of new DNA 3’-end to prime reverse transcriptase of element RNA – After 2nd strand synthesis, element has been replicated at target site • New round of transposition begins when the LINE is transcribed • LINE polyadenylation signal is weak, so transcription of a LINE often includes exons of downstream host DNA 23-32 Nonautonomous Retrotransposons • Nonautonomous retrotransposons include very abundant human Alu elements and similar elements in other vertebrates • Cannot transpose by themselves as they do not encode any proteins • Take advantage of retrotransposition machinery of other elements such as LINE • Processed pseudogenes likely arose in same manner 23-33 Group II Introns • Group II introns – Retrohome to intronless copies same gene by: • Insertion of an RNA intron into the gene • Followed by reverse transcription • Then second-strand synthesis – Retrotranspose by: • Insertion of an RNA intron into an unrelated gene • Target-primed reverse transcription • Lagging-strand DNA fragments as primers • Group II retrotransposition: – Forerunner of eukaryotic spliceosomal introns – Accounted for appearance in higher eukaryotes 23-34