* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download II. Transposable Elements in Bacteria Transposable Elements are
Gene desert wikipedia , lookup
Primary transcript wikipedia , lookup
Nucleic acid double helix wikipedia , lookup
X-inactivation wikipedia , lookup
Oncogenomics wikipedia , lookup
DNA supercoil wikipedia , lookup
Cell-free fetal DNA wikipedia , lookup
DNA vaccination wikipedia , lookup
Mitochondrial DNA wikipedia , lookup
Zinc finger nuclease wikipedia , lookup
Genome (book) wikipedia , lookup
Molecular cloning wikipedia , lookup
Genetic engineering wikipedia , lookup
Gene expression programming wikipedia , lookup
Short interspersed nuclear elements (SINEs) wikipedia , lookup
Homologous recombination wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Microsatellite wikipedia , lookup
Designer baby wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Metagenomics wikipedia , lookup
Minimal genome wikipedia , lookup
Human genome wikipedia , lookup
Point mutation wikipedia , lookup
Pathogenomics wikipedia , lookup
Microevolution wikipedia , lookup
Genomic library wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Extrachromosomal DNA wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
History of genetic engineering wikipedia , lookup
Non-coding DNA wikipedia , lookup
Genome editing wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Genome evolution wikipedia , lookup
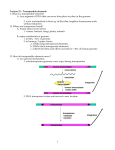
II. Transposable Elements in Bacteria Transposable Elements are DNA sequences that are capable of mediating their own movement (transposition) to new locations within the genome they inhabit. Barabara McClintock was awarded a Nobel Prize for her pioneering discovery of transposable elements in the genome of maize. Transposable elements of various types are widespread in genomes of eukaryotes and bacteria. In bacteria, transposable elements can generally be assigned to one of two major types, "Insertion Sequences (IS)" and "Composite Transposons". In practice, composite transposons are typically referred to simply as "transposons". Insertion sequences (IS's) are transposable elements whose only genes are directly related to promotion and regulation of their transposition, typically the gene for the so-called transposase enzyme. IS elements are between 700 - 2,000 bp in length and are characterized by short, terminal, inverted repeat sequences with the ORF or ORF's in between. They are normal constituents of many bacterial chromosomes and plasmids. Composite transposons generally consist of two copies of the same IS element flanking variable amounts of other DNA sequences coding for one or several genes with diverse functions. The best known transposons are those which were discovered as parts of antibiotic resistance plasmids.s The diagram below compares the typical structure of an IS element with the transposon Tn5. Tn5 carries 3 antibiotic resistance genes sandwiched between 2 copies of IS50. Note that the R Streptomycin-resistance gene is inactivated by a mutation, so they will not make strains Str . Only IS elements will be discussed below. IS elements participate in rather bewildering array of molecular events that alter the genomes which they inhabit. The most important of these are: Transposition IS movement and insertion at a different location in the same DNA molecule, or in different molecule in the cell. The transposition process is often accompanied by replication of the IS (replicative transposition), leading to an increase in the copy # of the IS. The diagram below shows replicative transposition of an IS. Insertional Inactivation Insertion of an IS within a coding sequence generally leads to the loss of gene function (null mutation). Homologous Recombination Multiple copies of the same IS in the same cell are substrates for homologous recombination events that may lead to DNA deletions, sequence inversions, or fusion of separate DNA molecules. For example, homologous recombination between copies of the same IS element in a conjugal plasmid and the bacterial chromosome leads to formation of Hfr strains, as shown below. IS F+ IS IS Hfr IS The multiplicity of transpositional and recombinational events associated with IS elements allows them to unlock the Pandora's box of genome plasticity for bacterial chromosomes and plasmids in which they are found. In fact, the K-12 laboratory strains of E. coli show considerable variability in the number and location of IS elements in their genomes due to transposition events that have occurred since the parent strain was first isolated in 1922.