* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Slide 1
DNA vaccination wikipedia , lookup
Nucleic acid tertiary structure wikipedia , lookup
Point mutation wikipedia , lookup
Nucleic acid double helix wikipedia , lookup
Molecular cloning wikipedia , lookup
Extrachromosomal DNA wikipedia , lookup
Cell-free fetal DNA wikipedia , lookup
RNA interference wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
History of genetic engineering wikipedia , lookup
DNA supercoil wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
History of RNA biology wikipedia , lookup
Non-coding DNA wikipedia , lookup
Epigenetic clock wikipedia , lookup
Designer baby wikipedia , lookup
Non-coding RNA wikipedia , lookup
Microevolution wikipedia , lookup
Epitranscriptome wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
DNA methylation wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Genomic imprinting wikipedia , lookup
RNA silencing wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Transgenerational epigenetic inheritance wikipedia , lookup
Epigenetics of depression wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Primary transcript wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Behavioral epigenetics wikipedia , lookup
Bisulfite sequencing wikipedia , lookup
X-inactivation wikipedia , lookup
Cancer epigenetics wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Neocentromere wikipedia , lookup
Epigenetics wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Histone acetyltransferase wikipedia , lookup
Epigenetics in stem-cell differentiation wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Nutriepigenomics wikipedia , lookup
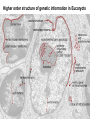
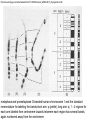
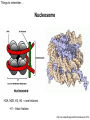
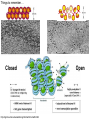
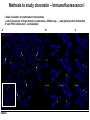
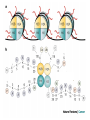
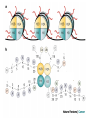
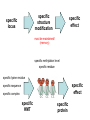
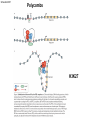
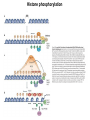
Přírodovědecká fakulta UK EPIGENETIKA MB150P85 Petr Svoboda mail: tel: [email protected] 241063147 A few comments on the course: - the goal of this course is to present you the latest original information on epigenetics, to give you some idea on how is such information obtained and to make you a better scientist. - this course is designed for advanced students, particularly for those who consider career in science. The course is modeled after advanced Msc./PhD. courses at the University of Pennsylvania and is likely very different from anything you have experienced at the university, so far. - this course requires active participation and is quite is demanding. You will have to read the original literature, make a few homeworks, take an exam and write a scientific text. You earn your credits. - the course is taught in English now. Mp3 records in Czech from 2007 Spring semester are available per request. - there are no stupid question. It is stupid not to ask. Exception: don’t ask if this or that is going to be included in the exam because the answer is always YES. - take the course as a challenge. Don’t take it if you don’t like to be challenged. Resources Suggested reading: Allis et al., Epigenetics Alberts et al. Molecular Biology of the Cell Passarge E., Color Atlas of Genetics Tost, Epigenetics Original articles a reviews will be provided during the course (as .pdf ) Resources Suggested reading for minimalists: NCBI - literature (Pubmed, OMIM), sequences (Genbank) a BLAST Ensembl - anotated sequences, data mining BioGPS – atlas of gene expression BCM Search Launcher – sequence analysis (old and decaying) Google and Wikipedia work very well too Additional information Office hours: - no specific time (unless you insist), you can come anytime - to make sure I’ll have time, please, write me or call me in advance Course requirements and the final exam - “take-home” two week exam - the code of academic integrity will be strictly enforced EPIGENETIKA B150P85 Introduction -overview of the course, basic concepts of epigenetic marks, diversity of epigenetic mechanisms and effects Lecture 1 24.2. 2011 Histones I - concept of chromatin structure. Heterochromatin and euchromatin. Core histones, linker histones, replacement histones, protamines. Methods for studying chromatin. Histones II - histone modifications, polycomb proteins, acetylation, fosforylation and histone methylations, effects on gene expression. DNA methylation I Lecture 2 10.3. 2011 - molecular basis of DNA methylation. CpG and non-CpG methylation. Adenosin methylation. Metods for studying DNA methylation. Bisulfite sequencing. DNA methylation II - effects of DNA methylation on gene expression, Methyl-binding proteins and mechanisms of inhibition of gene expression, distribution of DNA methylation within genes and mammalian genomes. RNA silencing I – molecular machines for RNA silencing Lecture 3 24.3. 2011 -“historical” introduction into RNA silencing. Post-transcriptional effects. Roles and effects of dsRNA. Proteins and complexes in RNA silencing. RNA silencing II - RNAi technology - experimental and therapeutic use. Design of RNAi experiments RNA silencing III – roles of RNA silencing pathways -miRNA pathway, chromatin connection. Imprinting Lecture 4 7.4. 2011 - concept of imprinting, mammalian imprinting. Molecular mechanisms of imprinting. Role of imprinting, Battle of the sexes. X-inactivation - principles and different strategies for dosage compensation. Control of X-inactivation in mammals. Epigenetic reprogramming in mammalian life-cycle Lecture 5 21.4. 2011 -integration of epigenetic modification in the mammalian life cycle. Reprogramming of gene expression during development, artificial reprogramming – the traditional view. Chromatin in transcribed regions - journal club Integrated view of epigenetic regulation of gene expression Lecture 6 5.5. 2011 - establishment of pluripotency in ES cells and embryos Course overview, feedback session EPIGENETICS Epigenetics deals with heritable information which is not encoded in the DNA sequence Such information can be encoded in: • structure and chromatin modifications • DNA modifications • RNA molecules HISTONES I (CHROMATIN STRUCTURE AND FUNCTION) Regulation of complex genomes is a problem TF binding site length? Core promoter length? Homo sapiens E. coli 42= 16 43= 64 44= 256 45= 1024 46= 4096 47= 16384 48= 65356 chromatin represents a structural solution for maintaining and accessing complex genomes Higher order structure of genetic information in Eucaryots HETEROCHROMATIN vs. EUCHROMATIN Dyes, like carminic acetic acid or orceine can be used to stain certain domains of a chromosome. The resulting pattern is characteristic for the respective chromosome of a species. During interphase, the chromosomal structure is usually resolved. The intensity of the nuclear staining becomes feebler and less uniform than that of the chromosomes. The stainable substance has been called chromatin by E. HEITZ (formerly at the Botanical Institute of the University of Hamburg, 1927, 1929). He distinguished between heterochromatin and euchromatin. Heterochromatin are all the intensely stained domains, euchromatin the diffuse ones. Heterochromatin is usually spread over the whole nucleus and has a granular appearance. It is known today that the heterochromatic domains are those where the DNA is tightly packed (strongly condensed) which is the reason for their more intense staining. The euchromatic domains are less tightly packed. http://www.biologie.uni-hamburg.de/b-online/e11/11c.htm#05 CHROMOSOME BANDING TECHNIQUES Prior to 1960, when Moorehead and Nowell described the use of Giemsa in their chromosome preparations, conventional cytologic stains such as acetoorcein, acetocarmine, gentian violet, hematoxylin, Leishman's, Wright's, and Feulgen stains were used to stain chromosomes. The Romanovsky dyes (which include Giemsa, Leishman's, and Wright's stain) are now recommended for conventional staining, because the slides can be easily destained and banded by most banding procedures. Orcein-stained chromosomes cannot be destained and banded; therefore, orcein is generally not used in routine chromosome staining. Giemsa stain is now the most popular stain for chromosome analysis (Gustashaw, 1991). Banding protocols http://homepage.mac.com/wildlifeweb/cyto/text/Banding.html http://www-biology.ucsd.edu/classes/bimm110.SP06/lectures_WEB/L08.05_Cytogenetics.htm http://www-biology.ucsd.edu/classes/bimm110.SP06/lectures_WEB/L08.05_Cytogenetics.htm metaphase and prometaphase G-banded human chromosome 1 and the standard nomenclature for labeling the bands;short arm: p (petite); long arm: q; 1 - 4 regions for each arm labeled from centromere towards telomere each region has several bands, again numbered away from the centromere http://fig.cox.miami.edu/~cmallery/150/proceuc/chromosome.jpg Evolution of chromatin structure models Molecular Biology of the Cell 1994 Molecular Biology of the Cell 2002 Molecular Biology of the Cell 2007 Things to remember … Nucleosome H2A, H2B, H3, H4 – core histones H1 – linker histone http://en.wikipedia.org/wiki/File:Nucleosome.JPG Things to remember … Closed http://sgi.bls.umkc.edu/waterborg/chromat/chroma09.html Open Things to remember … apparent global chromatin patterns are underlied by repetitive sequences Martens 2005 NUCLEOSOME AND CORE HISTONES H2A, H2B, H3, H4 – core histones H1 – linker histone Replication-dependent core histones - localized in large clusters (common chromatin domains? RNA processing?) - the major human cluster - 6p21(mouse chr. 13) - smaller clusters on 1p21 (mouse chr. 3) and 1q42 (mouse chr. 11) - the major cluster tends to colocalize with Cajal bodies (functional link isn‘t well understood) histone type cluster gene nomenclature HIST1H2AG family member older nomenclature and synonyms can be clarified at the GNF Symatlas and NCBI webpages Marzluff 2002 Expression of core histones cell-cycle dependent http://www.unc.edu/depts/marzluff/research.html specific 3‘ end processing CPSF-73 http://www.reactome.org/cgi-bin/eventbrowser?DB=gk_current&ID=77588& Mammalian core histone variants H2A.X - estimated to make 10% of nuclear H2A in mammals - rapidly phosphorylated in a response to DNA damage CENP-A (variant of histone 3, Cid in Drosophila) - found at centromeric regions macroH2A - enriched on the inactive X chromosome H2A.Z - possibly involved in initial steps of gene activation in euchromatin H3.3 - deposited within chromatin independent on DNA replication - enriched at sites of transcription - accumulates in non-cycling cells H3.1 - synthesized and deposited during S-phase H2A.Bbd - excluded from the inactive X chromosome - H2A.Bbd histone octamer organizes only approximately 130 bp of DNA Protamines - non-histone replacement in sperms Braun 2001 Methods to study chromatin – Immunofluorescence I - Small resolution on mammalian chromosomes - useful of analysis of large domains (centomeres, rDNA arrays …) and global protein distribution -IF and FISH combination - colokalization A B 349 CENP-A 349 350 HA UBF UBF 349 Merge Merge Merge HEK293 C Methodsto study chromatin – Immunofluorescence I - Higher resolution in polytene chromosomes in Drosophila Polytene chromosomes (blue) stained for Hairy (green) and Groucho (red) binding Methods to study chromatin – Chromatin IP • good resolution (typically 0.5 - 1.0 kb) • useful for analysis of individual genes, promoters • genome-scale analysis nowadays possible • relatively expensive tips and tricks Detection: •qPCR •promoter/tiling microarray = ChIP-Chip •deep sequencing = ChIP-Seq Methods to study chromatin – Chromatin IP human rDNA repeat A 5´ETS 10kb 5kb 0kb 18S 5.8S 1kb 3kb 6kb 15kb 20kb 25kb 30kb 35kb 40kb 43kb 28S 13kb 20kb 29kb 38kb 42kb B 14 12 349 % of input 10 unspecific antibody 8 6 4 2 0 GAPDH 1kb 3kb 6kb 13kb 20kb 29kb 38kb 42kb HISTONES II (MODIFICATIONS AND THEIR INTERPRETATION) http://biology.plosjournals.org/perlserv?request=get-document&doi=10.1371/journal.pbio.0020136 http://sgi.bls.umkc.edu/waterborg/chromat/chroma09.html Proc Natl Acad Sci U S A. 1964 May; 51(5): 786–794. http://biology.plosjournals.org/perlserv?request=get-document&doi=10.1371/journal.pbio.0020136 Kuo 1998 Histone acetylation - deacetylation Kuo 1998 Histone acetylation - deacetylation Kuo 1998 Histone acetylation - deacetylation Histone acetylation Deposition-related (B HATs) Transcription-related (A HATs) Annemieke 2003 Histone deacetylases Trichostatin A is an inhibitor of histone deacetylases. + Sir2 family of deacetylases - target nonhistone proteins Histone methylation - lysine residues SET domain HMTs http://www.imt.uni-marburg.de/bauer/research.html Bannister 2002 specific structure modification specific locus specific effect must be maintained! (memory) specific methylation level specific residue specific lysine residue specific effect specific sequence specific complex specific HMT specific protein Histone methylation - lysine residues Bannister 2002 histone code concept Shi 2007 Histone methylation is reversible! JmjC domain (JumanjiC) Histone methylation - arginine residues http://www.imt.uni-marburg.de/bauer/research.html Abcam = common source of Abs and information http://www.abcam.com/ Bannister 2005 Shi 2007 mono di tri and back Arney 2007 … BACK TO HETEROCHROMATIN vs. EUCHROMATIN Martens 2005 … BACK TO HETEROCHROMATIN vs. EUCHROMATIN DISTINCT REGIONS WITHIN CHROMATIN Histone code and its interpretation Bannister 2002 Histone code and its interpretation Complexes, complexes, complexes … Bannister 2002 Li 2002 Schwartz 2007 Polycombs Schwartz 2007 Polycombs H3K27 Histone phosphorylation It has been estimated that a typical human cell must repair over 10,000 DNA lesions per day (Lindahl, T. Nature, 1993). Histone phosphorylation