* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Gene Set Enrichment Analysis
Gene therapy of the human retina wikipedia , lookup
Short interspersed nuclear elements (SINEs) wikipedia , lookup
Neuronal ceroid lipofuscinosis wikipedia , lookup
Transposable element wikipedia , lookup
Copy-number variation wikipedia , lookup
Genetic engineering wikipedia , lookup
Epigenetics in learning and memory wikipedia , lookup
Oncogenomics wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Gene therapy wikipedia , lookup
X-inactivation wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Pathogenomics wikipedia , lookup
Quantitative trait locus wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Gene nomenclature wikipedia , lookup
Essential gene wikipedia , lookup
Public health genomics wikipedia , lookup
History of genetic engineering wikipedia , lookup
Gene desert wikipedia , lookup
The Selfish Gene wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Genome evolution wikipedia , lookup
Genomic imprinting wikipedia , lookup
Minimal genome wikipedia , lookup
Gene expression programming wikipedia , lookup
Ridge (biology) wikipedia , lookup
Biology and consumer behaviour wikipedia , lookup
Microevolution wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Genome (book) wikipedia , lookup
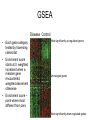
Gene Set Enrichment Analysis GSEA: Key Features • Ranks all genes on array based on their differential expression • Identifies gene sets whose member genes are clustered either towards top or bottom of the ranked list (i.e. up- or down regulated) • Enrichment score calculated for each category • Permutation test to identify significantly enriched categories • Extensive gene sets provided via MolSig DB – GO, chromosome location, KEGG pathways, transcription factor or microRNA target genes GSEA Disease Control • Each gene category tested by traversing ranked list • Enrichment score starts at 0, weighted increment when a member gene encountered, weighted decrement otherwise Most significantly up-regulated genes Unchanged genes • Enrichment score – point where most different from zero Most significantly down-regulated genes Enrichment Score • Enrichment Score (ES) is calculated by evaluating the fractions of genes in S (‘‘hits’’) weighted by their correlation and the fractions of genes not in S (‘‘misses’’) present up to a given position i in the ranked gene list, L, where N genes are ordered according to the correlation, 4 GSEA algorithm GSEA: Permutation Test • Randomise data (groups), rank genes again and repeat test 1000 times • Null distribution of 1000 ES for geneset Null distribution of enrichment scores Actual ES • FDR q-value computed – corrected for gene set size and testing multiple gene sets