* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Loose Ends on Chapters 3,5,6
P-type ATPase wikipedia , lookup
Cell nucleus wikipedia , lookup
Phosphorylation wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Cytoplasmic streaming wikipedia , lookup
Protein moonlighting wikipedia , lookup
SNARE (protein) wikipedia , lookup
Protein phosphorylation wikipedia , lookup
Intrinsically disordered proteins wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Cytokinesis wikipedia , lookup
Circular dichroism wikipedia , lookup
Magnesium transporter wikipedia , lookup
Cell membrane wikipedia , lookup
Signal transduction wikipedia , lookup
Western blot wikipedia , lookup
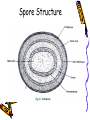
Loose Ends on Chapters 3,5,6 Summer Microbiology Spore survival • Dipicolinic acid and Ca++ account for 15% of the total spore mass • Dipicolinic acid theoretically may contribute to the stability of the nucleic acids which is a contributory to the spore’s survival- The Ca and the dipicolinic acid may enhance the activity of DNA binding proteins that are vital to the spore’s ability to resist radiation • Calcium contributes to the ability of the spore to resist destruction by oxidizing agents and dry heat – also steam Spore Structure Seven Steps and More in Spore Formation Revisited • Stress or unfavorable environmental conditions • Replication of DNA • Membrane begins to form to separate cells – Forespore septum begins to show the formation of the forespore • Membrane continues to grow and engulfs the DNA into the forespore Spore formation Continued • The cortex is laid down and Calcium and dipicolinic acid are accumulated here • Protein coats then form around the cortex • The exosporium and then the spore coat are made to surround the spore • At this point the spore structure is completed Spore Release • Lytic enzymes destroy the sporangium releasing the spore • This process takes about 23 hours in B. subtilis Flagellar Structure • The M Ring is anchored in the cel membrane of the bacterium. • A shaft that is attached to the hook and flagellum extends form the shaft. • In Gram Positive cells the S ring is attached to the cell wall and does not rotate • In Gram Negative cells, the P and L rings act as a bearing for the rotating flagellum Flagellar Motor Proteins • Mot A and Mot B • These form a proton channel for the establishment of a proton gradient • The Motor proteins also assist in the anchoring of the complex to cell wall peptidoglycans • Fli G is the motor protein that generates flagellar rotation Rotational symmetry of the C ring and a mechanism for the flagellar rotary motor • Dennis R. Thomas, David Gene Morgan, and David J. DeRosier • Proc Natl Acad Sci U S A. 1999 August 31; 96(18): 10134–10139. PMCID: 17855 Reference • The cytoplasmic component of the bacterial flagellar motor. • I H Khan, T S Reese, and S Khan • Proc Natl Acad Sci U S A. 1992 July 1; 89(13): 5956–5960 Chemoreceptors • MCP – Methyl accepting chemotaxis proteins • Localized in patches at the ends of the bacilli in E. coli • React to stimuli through a series of steps utlizing proteins Concepts • Conformation change in protein structure • Methylation of proteins • Phosphorylation of proteins Increase in nutrient binding • Environmental stimulus – nutrient molecule • Nutrient molecule in the gradient binds to the MCP protein( change in conformation) • Che A is dephosphorylated – loses a phosphate • Counterclockwise rotation occurs in the bacteria No nutrient detected in the environment • No nutrient binding • Che A is phosphorylated( gains a phosphate group) • Phosphate is then donated to Che Y • Interacts with Fli switch to causes clockwise rotation or tumbling – random and undirected Two Component Phosphorelay System • Two Component Phosphorelay System is similar in response to oxygen, light, hear and osmotic gradients. Active Transport • Movement against the concentration gradient from low concentration outside of the cell to higher inside the cyotplasm • Requires the input of energy to drive the reaction forward Active Transport and ATPBinding Cassette Transporters ABC Transporters • Large group of transporters • Two hydrophobic domains in the membrane and two nucleotide binding domains at the cytoplasmic surfaces • The membrane spanning portions form a pore • The nucleotide binding domains bind ATP for the hydrolysis of ATP to produce energy to drive molecules through the membrane Salmonella ABC transporter E. Coli and active transport • The sugars arabinose, maltose galactose, and ribose are transported by this mechanism in bacteria • Also amino acids may pass through the cell membrane in this manner Porins • Porins are channels located in the outer membrane of the Gram Negative bacteria Omp F Siderophore Active transport • Uniport – One ion or molecule moves against the concentration gradient • Symport – A concentration gradient established by an ion, drives solute transport of another molecule against the concentration gradient • Antiport – Sodium is pumped outward in response to an inward movement of protons Importance of this mechanism • The sodium pumped to the outside of the cell is also used in transport • It binds to the outside of a different transport protein. • When it binds it changes the shape of the protein • The protein is then able to bind to molecules to move them into the cell E. coli • Has multiple transport systems for many nutrients • The diversity of these transport system provides the ability to survive in diverse environments Group Translocation • A molecule is transported ito the cell while being chemically altered • This is energy dependent PTS • Best characterized system is the PTS • Phosphoenolpyruvate: sugar phosphotransferase system • Transports sugars while phosphorylating them Components • Two enzymes( EI and EII) • Low Molecular weight – heat stable protein( HPr) Steps • High energy Phosphate is transferred from phosphoenolpyruvate to EII with the help of EI and HPr • The sugar molecule to be transported is phosphorylated as it goes across the membrane by EII • EII is specific for the sugar that it transports PTS