* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download ERF/AP2 Subfamily A3 and ER/AP2 Subfamily A6 Genes
Zinc finger nuclease wikipedia , lookup
Non-coding DNA wikipedia , lookup
Pathogenomics wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
X-inactivation wikipedia , lookup
Oncogenomics wikipedia , lookup
Transposable element wikipedia , lookup
Genomic imprinting wikipedia , lookup
Public health genomics wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Epigenetics in learning and memory wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Genetically modified crops wikipedia , lookup
Copy-number variation wikipedia , lookup
Point mutation wikipedia , lookup
Neuronal ceroid lipofuscinosis wikipedia , lookup
Genome (book) wikipedia , lookup
Saethre–Chotzen syndrome wikipedia , lookup
Genetic engineering wikipedia , lookup
Genome evolution wikipedia , lookup
Genome editing wikipedia , lookup
Genomic library wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
The Selfish Gene wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Gene therapy wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Gene expression profiling wikipedia , lookup
Gene expression programming wikipedia , lookup
History of genetic engineering wikipedia , lookup
Gene desert wikipedia , lookup
Gene nomenclature wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Helitron (biology) wikipedia , lookup
Microevolution wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
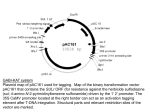
ERF/AP2 Subfamily A3 and ERF/AP2 Subfamily A6 Genes: AT1G64380 and AT4G39780 ERF/AP2 DREB Subfamily • Characterized by AP2 domain • AP2 family genes-shown to participate in regulation of embryo development • Encodes putative transcription factors (DNA binding motif) • ERF family: CBF and DREB subfamilies – DREB subfamilies contain AP2 and DREB motifs – DREB transcription factors control expression of drought-inducible genes Where is gene AT1G64380? 976 bp 5’ 3’ 742 bp AT1G64370 1,528 bp 2,247 bp AT1G64380 AT1G64385 7,861 bp What is gene AT1G64380’s structure? 185 bp UTR 5’ Coding Region (1,008 bp) UTR 335 bp Not part of gene 359 bp 3’ Where is the T-DNA insert located? LBb1 T-DNA insert: 300 bp RV 185 bp UTR 5’ Coding Region (1,008 bp) FW UTR 335 bp Predicted T-DNA insertion site: 1,206th bp of the gene WT expected length: 1,195 bp T-DNA expected length: 830 bp Not part of gene 359 bp 3’ Where is gene AT1G64380 expressed? Gene Chip Data Expression Levels Chalazal heart stage Chalazal seed coat/ linear cotyledon stage Chalazal seed coat/ Mature green stage 471.6 937.1 406.05 Floral bud/ reproductive Leaf/Vegetative 105.45 What are the plant genotypes? Homozygous T-DNA (4, 6, 9) 2 T-DNA bands: Top band matches expected amplified T-DNA size (830bp) Lower band approx. 750 bp Wild type Expected WT size: 1,195 bp What are the possible T-DNA configurations? LBb1 T-DNA insert LBb1 T-DNA insert: 300 bp RV 185 bp UTR Coding Region (1,008 bp) 5’ FW A concatamer! UTR 335 bp Not part of gene 359 bp 3’ What are the possible T-DNA configurations? LBb1 T-DNA insert: 300 bp RV 185 bp UTR 5’ Coding Region (1,008 bp) FW Two inserts located very close to each other; sequencing inconclusive UTR 335 bp T-DNA insert LBb1 Not part of gene 359 bp 3’ What do these results mean? Wildtype (WT/WT) Hemizygous (WT/T-DNA) Homozygous (T-DNA/T-DNA) 7 0 3 Presence of homozygous mutant plants: Knockout of gene AT1G64380 does NOT lead to seed lethality Where in Arabidopsis Thaliana is gene AT1G64380 transcribed? Use RT-PCR to detect AT1G64380 mRNA levels 04/28/09 //120 V // 1 hour // 1% agarose Tubulin control Genomic DNA water g DNA Silique -RT Siliqe +RT Leaf –RT Leaf +RT Ladder Gene AT1G64380 is expressed in both Arabidopsis leaf and silique tissues; supports Gene Chip data How can we visualize where gene AT1G64380 is transcribed? Promoter Cloning -no recombinant plasmids found in 42 bacteria colonies screened ASCI restriction digest and PCR colony screening Expected size of recombinant: 4.3 kb WHY? Initial ligation was not successful Negative control Positive control Pentr only PCR to amplify promoter region Pentr + gene Isolate plasmids from transformed E-coli No promoter region detected Observation of mutant siliques *Location of T-DNA insert! What is gene AT4G39780’s structure? 3’ 5’ orientation RV 256 bp UTR Coding Region (819 bp) 5’ 146 bp UTR 3’ FW Where is the T-DNA insert located? LBb1 T-DNA insert: 169 bp RV 256 bp UTR 5’ Coding Region (819 bp) FW Predicted T-DNA insertion site: 695th bp of the gene WT expected length: 741 bp T-DNA expected length: 569 bp 146 bp UTR 3’ Where is gene AT4G39780 expressed? Gene Chip Data Chalazal seed coat/ globular stage Floral Bud/ reproductive General Seed Coat/ Globular Stage General Seed coat/ Heart stage General seedcoat/ pre-globular stage Leaf/Vegetative What are the plant genotypes? All plants are wild type Expected WT size: 741 bp What do these results mean? Enough plants were screened T-DNA was not inserted into the gene Fatality of knockout to seed development is inconclusive No further analysis can be done on these plants Future Research AT1G64380 AT4G39780 *No phenotypic difference observed in mutant siliques •No T-DNA inserts found from screening 24 plants Look at Arabidopsis mutant leaves Check protein production levels Obtain different SALK lines for this gene Obtain a different SALK line for this gene --Repeat previous methods Acknowledgements Thank you to Anhthu, Kristen, Daisy, Brandon, Min, Tomo, Kelli, Ingrid and Dr. Goldberg for making this lab experience happen!! This has been an incredible quarter.