* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download PowerPoint 簡報
Short interspersed nuclear elements (SINEs) wikipedia , lookup
Behavioral epigenetics wikipedia , lookup
Epigenetics of depression wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Genetic engineering wikipedia , lookup
DNA supercoil wikipedia , lookup
Molecular cloning wikipedia , lookup
Genome evolution wikipedia , lookup
Bisulfite sequencing wikipedia , lookup
Zinc finger nuclease wikipedia , lookup
Protein moonlighting wikipedia , lookup
DNA vaccination wikipedia , lookup
Gene expression profiling wikipedia , lookup
Transcription factor wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Extrachromosomal DNA wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Non-coding DNA wikipedia , lookup
Epigenetics in stem-cell differentiation wikipedia , lookup
Epigenetics wikipedia , lookup
Point mutation wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Microevolution wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
History of genetic engineering wikipedia , lookup
Designer baby wikipedia , lookup
Cancer epigenetics wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Helitron (biology) wikipedia , lookup
Primary transcript wikipedia , lookup
Histone acetyltransferase wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Epigenomics wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Epigenetics in learning and memory wikipedia , lookup
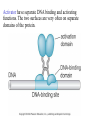
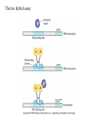
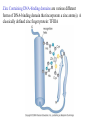
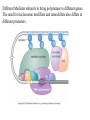
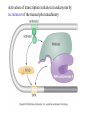
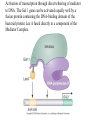
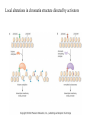
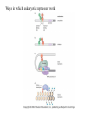
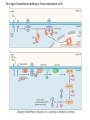
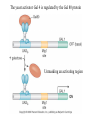
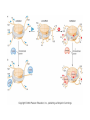
Chapter 17 Gene regulation in eukaryotes • Many eukaryotic genes have more regulatory binding sites and are controlled by more regulatory proteins than are typical bacterial genes. • An enhancer binds regulators responsible for activating the gene at the given time and place. • Insulators block activation of the promoter by activators bound at the enhancer. • A silencer binds regulators responsible for repressing the gene at the given time and place. Activator have separate DNA binding and activating functions. The two surfaces are very often on separate domains of the protein. Gal4 binds to four sites located 275 bp upstream of GAL1. When bound there, in the presence of galactose, Gal4 activates transcription of the GAL1 gene 1000-fold. Domain swap experiments A reporter gene consists of binding sites for the yeast activator inserted upstream of the promoter of a gene whose expression level is readily measured. Lex A, a bacterial repressor • In some cases, DNA binding domain and activating domains are even carried on separate polypeptides, and form a complex on DNA. • Herpes virus activator VP16 interacts with the Oct1 DNA-binding protein found in cells infected by this virus. The two hybrid assay Eukaryotic regulators use a range of DNAbinding domains, but DNA recognition involves the same principles as found in bacterial • There is no fundamental difference in the ways DNA-binding proteins from different organisms recognize their sites. • Eukaryotic regulators often bind as dimers and recognize specific DNA sequences using an αhelix inserted into the major groove. • Some regulators in eukaryotes bind DNA as heterodimers. Homeodomain is a class of helix-turn-helix DNA-binding domain and recognizes DNA in essentially the same way as those bacterial proteins. They were discovered in Drosophila where they control many basic developmental programs. Zinc Containing DNA-binding domains are various different forms of DNA-binding domain that incorporate a zinc atom(s). A classically defined zinc finger protein: TFIIIA Leucine zipper motif combines dimerization and DNA-binding surfaces within a single structural unit. Leucine-zipper-containing proteins often form heterodimers as well as homodimers. Helix-loop-helix proteins: The two helices are separated by a flexible loop that allows them to pack together. Leucine zipper and HLH proteins are often called basic zipper and basic HLH proteins: the region of the αhelix that binds DNA contains basic amino acid residues. Activating regions are not welldefined structures • Activating regions are grouped on the basis of amino acid content. • Acidic activating region: Gal 4 • Glutamine-rich activating region: Sp1 • Proline-rich activating region: CTF-1 Recruitment protein complexes to genes by eukaryotic activators • The activators recruit polymerase indirectly in two ways: the activator interacts with parts of the transcription machinery other than polymerase; activators can recruit nucleosome modifiers that alter chromatin in the vicinity of a gene and thereby help polymerase bind. • The eukaryotic transcriptional machinery contains numerous proteins in addition to RNA polymerase. • Many of these proteins come in preformed complexes such as the Mediator and the TFIID complex. In vivo, transcription Initiation requires additional Proteins, including the mediator complex • The high, regulated levels of transcription in vivo require the Mediator Complex, transcriptional regulatory proteins and in many cases, nucleosome-modifying enzymes. • Mediator is associated with the CTD “tail” of the large polymerase subunit through one surface, while presenting other surfaces for interaction with DNA-bound activators. Different Mediator subunits to bring polymerase to different genes. The need for nucleosome modifiers and remodellers also differs at different promoters. Mediator consists of many subunits, some conserved from yeast to human • There are various forms of Mediator, each containing subsets of Mediator subunits. • A complex consisting of Pol II, Mediator, and a some of the general transcription factors can be isolated from cells as a single complex in the absence of DNA---RNA Pol II holoenzyme. Activation of transcription initiation in eukaryotes by recruitment of the transcription machinery Activation of transcription through direct tethering of mediator to DNA. The Gal 1 gene can be activated equally well by a fusion protein containing the DNA-binding domain of the bacterial protein Lex A fused directly to a component of the Mediator Complex. Box 17-2 Chromatin Immunoprecipitation Recruitment can be visualized using ChIP Activators also recruit nucleosome modifiers that help the transcription machinery bind at the promoter • Recruitment of nucleosome modifiers can help activate a gene inaccessibly packaged within chromatin. • Histone acetyltransferases (HATs): add chemical groups to the tails of histone • ATP-dependent activity of SWI/SNF: remodel the nucleosomes Local alterations in chromatin structure directed by activators Action at a distance: Loops and Insulators • Many eukaryotic activators- particularly in higher eukaryotes- work from a distance. • IHF, an architectural protein, binds to DNA and by bending the DNA IHF helps the DNA-bound activator reach RNA polymerase at the promoter. • Specific elements called insulators control the actions of activators. Appropriate regulation of some groups of genes requires locus control regions • A group of regulatory elements collectively called the locus control region, or LCR, is found 30-50 kb upstream of the whole cluster of globin genes. • The simplest explanation is that regulatory proteins binds to the LCR and recruit chromatin modifying complexes to the region. Regulation by LCRs Signal integration and combinatorial control • Activators work together synergistically to integrate signals • When multiple activators work together, they do so synergistically. That is, the effect of two activators working together is greater (usually much greater) than the sum of each of them working alone. • Synergy can also result from activators helping each other bind under conditions where the binding of one depends on binding of the other. Cooperative binding of activators Signal integration • The HO gene, expressed only in mother cells when yeast divides by budding, is controlled by two regulators; one recruits nucleosome modifiers and the other recruits mediator. • If both activators are present and active, the action of SWI5 enables SBF to bind and activate the transcription of the HO gene. SWI5 can recruit nucleosome modifiers (histone acetyl transferases). SBF recruits Mediator Signal integration • Cooperative binding of activators at the human interferon gene • Viral infection triggers three activators: NFB, IRF, and jun/ATF. • The structure formed by these regulators bound to the enhancers is called an enhancersome. • HMG-1 has an architectural role in the process. The human -interferon enhancersome Combinatorial control lies at the heart of the complexity and diversity of eukaryotes Combinatorial control of the mating-type Genes from Saccharomyces cerevisiae Transcriptional repressors • Repressors can recruit nucleosome modifiers, they compact the chromatin or remove groups recognized by the transcriptional machinery, for example: histone deacetylases. Ways in which eukaryotic repressor work In the presence of glucose, Mig1 binds and switches off the Gal 1 genes. Mig1 recruits a “repressing complex” containing the Tup1 protein. Tup1 recruits histone deacetylase Signal transduction and the control of transcriptional regulators • Signals are often communicated to transcriptional regulators through signal transduction pathways • The initiating ligand is typically detected by a specific cell surface receptor. • The most common way in which information is passed through signal transduction pathways is via phosphorylation. Two signal transduction pathways from mammalian cells: Signal control the activators of eukaryotic transcriptional regulators in a variety of ways • In eukaryotes, transcriptional regulators are not typically controlled at the level of DNA binding (though there are exceptions). Regulators are instead usually controlled in one of two basic ways: 1. Unmasking an activating region 2. Transport into and out of the nucleus The yeast activator Gal 4 is regulated by the Gal 80 protein Unmasking an activating region Transport into and out of the nucleus Activators and repressors sometimes come in pieces • The activator can come in pieces: the DNA-binding domain and activating region can be on separate polypeptides. • Glucocorticoid receptor (GR) can either activate or repress transcription depending on the nature and arrangement of its DNA-binding sites at a given gene. • The terms” co-repressor” and “ co-activator ” are often applied to any auxiliary protein which is neither part of the transcriptional machinery nor itself a DNA-binding regulator, but which is nevertheless involved in transcriptional regulation. Gene “silencing” by modification of histone and DNA • Gene silencing is a position effect- a gene is silenced because of where it is located, not in response to a specific environmental signal. • The most common form of silencing is associated with a dense form of chromatin called heterochromatin; it appears dense compared to other chromatin. • Transcription can also be silenced by methylation of DNA by enzymes called DNA methylases. Silencing in yeast is mediated by deacetylation and methylation of histones: the final 1-5 kb of each chromosome is found in a folded, dense structure. SIR 2, 3 and 4 are silent information regulators, and Sir 2 is a histone deacetylase. Binds to the repeated sequences Rap1 recruits SIR2 Unacetylated tails then bind SIR3 and 4 Methylation of the tail of Histone H3 blocks the binding of the Sir2 • In high eukaryotes and in the yeast, silencing is typically associated with chromatin containing histones that are not only deacetylated but methylated as well. Histone modification and the histone code hypothesis • Histone code: different patterns of modifications on histone tails can be read to mean different things. • Multiple modifications at several positions in the histone tails are possible, the examples of H3 and H4, together with H2A and B. • Lysine 9 on the tail of histone H3: different modification states of this residue have different meaning. Acetylation of it is associated with actively transcribed genes. When lysine 9 is unmodified, it is associated with silenced regions. This lysine can be methylated then binds proteins that establish and maintain a heterochromatic state. DNA methylation is associated with silenced genes in mammalian cells Switching a gene off through DNA methylation and histone modification DNA-binding proteins (such as MeCP2) Then recruit histone deacetylases and histone methylases or chromatin remodeling complex •Imprinting: there are a few cases that one copy of a gene is expressed and the other is silent. Some states of gene expression are inherited through cell division even when the initiating signal is no longer present • The inheritance of gene expression patterns, in the absence of either mutation or the initiating signal, is called epigenetic regulation. • Nucleosome and DNA modifications can provide the basis for epigenetic inheritance. • Methylated nucleosomes in daughter molecules recruit protein bearing chromodomains to methylated the adjacent nucleosomes. •DNA methylation is reliably inherited, so-called maintenance methylases modify hemimethylated DNA. •Patterns of DNA methylation can be maintained through cell division