* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download mutation
Nutriepigenomics wikipedia , lookup
Primary transcript wikipedia , lookup
Epigenomics wikipedia , lookup
DNA supercoil wikipedia , lookup
Polymorphism (biology) wikipedia , lookup
Extrachromosomal DNA wikipedia , lookup
Human genome wikipedia , lookup
Genetic engineering wikipedia , lookup
SNP genotyping wikipedia , lookup
Genealogical DNA test wikipedia , lookup
Genome (book) wikipedia , lookup
Zinc finger nuclease wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
DNA damage theory of aging wikipedia , lookup
Genome evolution wikipedia , lookup
Genetic code wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Saethre–Chotzen syndrome wikipedia , lookup
History of genetic engineering wikipedia , lookup
Cancer epigenetics wikipedia , lookup
Designer baby wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Neuronal ceroid lipofuscinosis wikipedia , lookup
Koinophilia wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Cell-free fetal DNA wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Non-coding DNA wikipedia , lookup
Population genetics wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Genome editing wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Helitron (biology) wikipedia , lookup
Oncogenomics wikipedia , lookup
Microsatellite wikipedia , lookup
Microevolution wikipedia , lookup
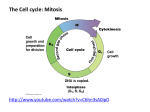
MUTATION AND POLYMORFISM Febr.19. 2016 Genetic variability - is increased by – mutation – sexual reproduction meiosis (generation of gametes) - homologous recombination (crossing over) - independent assortment of homologous chromosomes fertilisation - significance DNA Mutation (causing any change) DNA variants, alleles (any coding or non-coding sequence) Genetic (DNA) polymorphism Significance of mutation (for all species) Without mutation, evolution would not be possible. This is because mutations provide the "raw material" upon which the mechanisms of natural selection can act. Regarding the variants…. • normal or wild variant (allele) is the most frequent in a population • polymorphism (or polymorphic) is the variant (allele) if its frequency is › 1 % in the population (having no effect on phenotype) formerly: • mutation (or mutant) is the variant (allele) (formerly: if its frequency is ‹ 1 % in the population / disease causing, it has a negative connotation) Human Genom Variation Society offered: „sequence variant„ or vagy „allelic variant” Mutation: somatic mutations or germline mutations (recent) Polymorphism: in a population when the frequency is known Long way from mutation to polymorphism Appearance of new variant by mutation Survival of rare allele Increase in allele frequency after population expand New allele is fixed in population as novel polymorphism Classification of mutation types • by the cause • by the site • by the function • by the fitness • by the size By the cause mutations may be • Spontaneous • Induced Spontaneous mutation - Spontaneous chemical reactions in bases • Tautomerization • Depurination • Deamination - Errors in DNA related processes • Replication • Recombination • Repair E.g. Tautomers of adenine Frequent Rare Amino group Imino group T-A (Purine-Purine) results Depurination (hydrolysis) Deamination Mutation hot spot Repaired Not repaired DNA methylation (regulation of DNA functions, see epigenetics) Induced mutation Some environmental agent = mutagen – physical - radiation • Heat • UV • Ionizing – chemicals • Natural toxins • Synthetic substances – Laboratory substances – Pollutants – Chemoterapeutics Natural substances Psoralen Aflatoxin Aspergillus sp. Laboratory chemicals: acridine orange, Fluorescent dyes ethidium bromide, propidium jodide BrdU - thymine analogue A senescent endothelial cell stained with the fluorescent dye acridine orange to visualise the lysosomes. Acrylamide Polyacrylamide Pollutants E.g. benzpyrene polyaromatic hydrocarbons (PAH) Metabolized to epoxides in liver DNA adduct Mutation Biological warfare agent Mustard gase Iranian victim (end of 20th century) I. World war victim (beginning of 20th century) Correction of DNA errors • DNA polymerase with proofreading ability • Repair mechanisms are limited tothe nuclear but not mitochondrial DNA DNA repair mechanisms • Direct repair • Excision repair the change is reversed template is needed no template is needed mainly in prokaryotes in eukaryotes Repair of single strand damage (complementer strand is used as template) Xeroderma pigmentosum is caused by the defective nucleotide excision repair enzymes Repair of double strand breaks (DSB) may result loss of nucleotides = deleterious Sister chromatid (after S phase) or like in meiosis, homologous chromosome is used as template = safety Multicellular cell cycle M-phase cytokinesis mitosis M G2 Checkpoints: Restriction point G2 M (spindle) G1 G2 Restriction point S Interphase Go - Growth factors - anchorange Function and activity of checkpoint machinery G1 G2 DNA damage not complete DNA replication M free kinetochors sensor protein kinases transducer effector s t o p of c e l l repair c y c l e Ataxia telangiectasia (ATM=sensor) Its mutation causes rare, neurodegenerative, inherited disease (AR), that affects many parts of the body and causes severe disability, characterized by radiosensitivity and different tumors. Role of BRCA (transducer) proteins in DNA repair BRCA mutations are found in breast and ovarian tumors. p53 Site of mutations - in the organism • Somatic - in somatic cells localized- inherited within cells of an organism (mosaicism: tumors, antibody diversity, etc.) higher in dividing cells • Generative – in primordial germ line inherited from one generation to the next one And nondisjunction of sex chromosomes Site of mutations - in a gene may be 1 2 UTR UTR 3 4 1/ Promoter mutations decreased transcription 2/ Exon mutations amino acid change or truncated protein (stop) see later 3/ Intron mutations errors in splicing 4/ Polyadenylation site mutations decreased mRNA stability 5 5 UTR disturbed ribosome binding Mutations of other regulatory sequences (enhancers, silencers) also may influence transcription. B.R. Korf: Human Genetics and Genomics,2006 Splicing mutations B.R. Korf: Human Genetics and Genomics,2006 Splicing mutations B.R. Korf: Human Genetics and Genomics,2006 Different mutations of a gene may lead to different malfunctions of the protein (=CFTR) Most frequent Function and mutations Back mutation or reversion is a point mutation that restores the original sequence and hence the original phenotype. Lethal mutations are mutations that lead to the death of the organisms which carry the mutations. Gain-of-function mutations - change the gene product such that it gains a new and abnormal function. These mutations usually have dominant phenotypes. Loss-of-function mutations - gene product having less or no function. Phenotypes associated with such mutations are most often recessive. Exception is when the reduced dosage of a normal gene product is not enough for a normal phenotye (this is called haploinsufficiency). Dominant negative mutations - the altered gene product acts antagonistically to the wild-type allele. These mutations are characterised by a dominant phenotype. In humans, dominant negative mutations have been implicated in cancer (e.g. mutations in genes p53, ATM). Fitness and mutations - Most are neutral – during evolution later may be harmful or beneficial - Some are beneficial – - harmful one mutates back to wild - getting beneficial function – diversity of antibody - CCR532 – HIV resistency - sickle cell anemia – malaria resistency - Some are harmful – causing diseases (all monogenic inherited diseases) Size of mutations • Large Genome mutation = change of chromosome number Cytogenetics • Medium Chromosome mutations = change of chromosome structure • Small gene mutations = ranging from a change of single nucleotide to a whole gene (not visible) Affecting the lenght of DNA Deletion (single base or shorter-longer sequences) Insertion (single base or shorter-longer sequencesrepetitive more insertion than deletion No effect on the length of DNA nucleotide substitution Repetitive insertions – Tandem repeats • Satellite DNA – pericentromeric heterochromatin • Minisatellite (VNTR) – 10-60 bp – Telomere • Microsatellite (STR=short tandem repeats) – 2- some bp – good markers of kinship – Repeat number expansion diseases – Interspersed repeats • SINEs (Short Interspersed Elements), • LINEs (Long …) e. g. L1 38 Microsatellite (STR = short tandem repeats) • 1-4 bp • Trinucleotide (triplet) repeats are very frequent – only few of them cause disease Trinucleotide repeats may be either in coding(C) or noncoding (NC) region NC C NC (huntingtin) (Huntingtin) C C coding NC noncoding Polyglutamine disorders • CAG repeats • Neurodegenerative disorders • Different proteins • Gain of function mutations • Variable length • Expansion • Replicational slippage Polyalanine disorders Replication slippage Huntingtin MATLEKLMKAFESLKSFQQQQQQQQQQQQQQQQQQQQQQQPPPP PPPPPPPQLPQPPPQAQPLLPQPQPPPPPPPPPPGPAVAEEPLHRPK KELSATKKDRVNHCLTICENIVAQSVRNSPEFQKLLGIAHELFLLCSDD... • • • • 350 kD protein ubiquitously expressed function unknown correlation between repeat size and age of onset and the severity of disease (Huntington chorea) Huntington healthy Polyglutamine disorders • CAG repeats Polyalanine disorders • GCX repeats • Neurodegenerative disorders •Developmental abnormalities • Different proteins • Transcription factors • Gain of function mutations • Loss of function mutations • Variable length • Constant length • Expansion • Stable • Replicational slippage • Uneven crossing over Uneven crossing over Uneven sister chromatid exchange Polyalanine disorder Disorder • Holoprosencephaly Gene ZIC2 Deletion or insertion of a single nucleotide (InDel) It is a frameshift mutation if number of nucleotide is not a multiple of three, and in-frame if number of nucleotide is a multiple of three DNA mRNA protein Mutant protein Medium InDel mutations • Deletion – Pl. Hypodontia (Deletion of Pax 9) • Insertion – (retro)transposons • Eg. L1 hemophilia A L1 is a LINE: Long Interspersed Elements L1 insertion and recombination in Hemophilia A VIII. Blood cloting factor gene (F8) Hemophilia A inversion mutation due to recombination between L1 repetitive sequences within (gray arrow) an outside (red arrow) the F8 gene 51 Single nucleotide substitution Nucleotide substitutions in coding region Transition Pyr ↔ Pyr Pu ↔ Pu More frequent in human Transversion Synonymous No change in amino acid Pyr ↔ Pu not synonymous change of amino acid or no amino acid Missense mutation – sickle cell anemia Frequences of disease causing mutations Polymorphism • Polymorphism appears at different levels: – Phenotype polymorphism – Protein polymorphism (Immunoglobulins, ABO blood groups) – Genetic (DNA) polymorphism Rate of genetic polymorphism – Identity between individuals = 99,5% – Difference between individuals = 0,5% DNA Polymorphism It is variation of DNA sequence that is common in given population Most are neutral, but some confer susceptibility or resistance to disease In human genome there are many, that is why can be used for personal identification Detection technics are available Genetic polymorphism • chromosomal (minor variants) • tandem repeats Satellite DNA pericentromeric heterochromatin Minisatellite (VNTR) Telomere Microsatellite (STR=short tandem repeats) • Single nucleotide polymorphism (SNP) Chromosomal polymorphism 1, 9,16 chromosomes centromeres Y chromosome long arm Nucleotide substitution=SNP (single nucleotide polymorphism) ATGGTAAGCCTGAGCTGACTTAGCGT ATGGTAAACCTGAGTTGACTTAGCGT SNP SNP Disease resistant population Disease susceptible population Genotype all individuals for thousands of SNPs ATGATTATAG geneX ATGTTTATAG Resistant people all have an ‘A’ at position 4 in geneX, while susceptible people have a ‘T’ Some disease associated SNPs Association ≠ correlation PPARG = Peroxisome Proliferator Activator Receptor γ, APOE = Apolipoprotein E; F5 = Factor V; CTLA4 = Cytotoxic T-lymphocyte Antigene 4; GSTM1 =Glutathione S-transferase Mu 1; INS = insulin; KCNJ11 = ATP sensitive K+channel coding gene; HF1/CFH = Complement factor H; COL1A1 = Collagen type A1; CARD15 = Caspase Recruitment Domain 15; CNV = copy number variations 1 kb - 5 megab About 1500 CNV 12% of genome 2900 gene is based on mutations and polymorphisms Giude to point mutations