* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download 4132010
Genetic engineering wikipedia , lookup
History of RNA biology wikipedia , lookup
Short interspersed nuclear elements (SINEs) wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Essential gene wikipedia , lookup
Non-coding DNA wikipedia , lookup
Epitranscriptome wikipedia , lookup
X-inactivation wikipedia , lookup
Human genome wikipedia , lookup
Gene nomenclature wikipedia , lookup
Pathogenomics wikipedia , lookup
Genomic library wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Transposable element wikipedia , lookup
Genomic imprinting wikipedia , lookup
Gene therapy wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Gene desert wikipedia , lookup
Gene expression programming wikipedia , lookup
Ridge (biology) wikipedia , lookup
Non-coding RNA wikipedia , lookup
Public health genomics wikipedia , lookup
Oncogenomics wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Biology and consumer behaviour wikipedia , lookup
Primary transcript wikipedia , lookup
Mir-92 microRNA precursor family wikipedia , lookup
Genome evolution wikipedia , lookup
Minimal genome wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
History of genetic engineering wikipedia , lookup
Genome (book) wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Gene expression profiling wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Microevolution wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Designer baby wikipedia , lookup
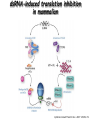
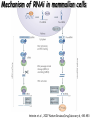
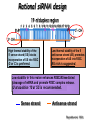
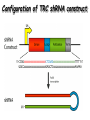
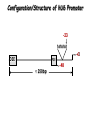
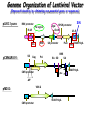
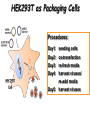
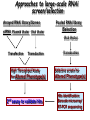
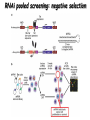
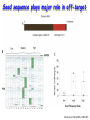
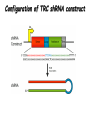
Resources and applications of TRC RNAi reagents in National RNAi Core Facility Lecturer: 林志隆 ( IMB&RNAi Core) 04/13/2010 The Nobel Prize in physiology /medicine 2006 Goes to Professor Andrew Z Fire, Ph.D. Stanford University School of Medicine Stanford, CA. USA Professor Craig C. Mello, Ph.D. University of Massachusetts Medical School Worcester, MA, USA RNAi: A gene silencing by dsRNA RNA interference (RNAi) A form of post-transcriptional gene silencing, mimicking the effect of loss-of-gene-function. RNAi does not result in stable genetic changes; but in lower animal or plants, RNAi effects can be inherited for one or two generations. Timeline of RNAi achievements Adapted from: http://www.invitrogen.com/etc/medialib/en/images/mainbody/ Thing/Data/Diagram.Par.12505.Image.-1.0.1.gif Dr. R Jorgensen’s Experiment C Napoli, C Lemieux, and R Jorgensen Plant Cell. 1990 April; 2(4): 279. • Attempts to overexpress chalcone synthase by inserting multiple copies of that gene into the plant’s genome. • Purple plants should become purpler... • Co-suppression: both endogenous and introduced genes silenced. • PTGS – but what is the causative factor? PTGS= Post-Transcriptional Gene Silencing PTGS in plants is due to small dsRNA dsRNA hypothesis explained this phenomenon Andrew J. Hamilton and David C. Baulcombe Science 1999 286: 950-52 Nature 391, 806-811 (19 February 1998) Long dsRNAs trigger non-specific silencing in mammalian dsRNA C. elegans Drosophila In mammal? Long dsRNA initiate IFN response in mammalian ---global gene silencing dsRNA-induced translation inhibition in mammalian Cytokine Growth Factor Rev. 2007 18:363-71. How to Apply RNAi to Mammalian System ? Effector of RNAi 長雙股RNA (long dsRNA) cytosol Gregory Hannon identified the “Dicer” – an enzyme that chops double-stranded RNA into little pieces. small-interfering RNA; siRNA - Length of siRNA: 21 nts to 23 nts. Nature. 2000;404:293-6; Nature. 2001;409:363-6 Nature 411, 494-498 (24 May 2001) What Does siRNA Do RISC: RNAi-induced silencing complex Guide/ antisense strand http://www.nature.com/focus/rnai/animations/index.html http://www.pbs.org/wgbh/nova/sciencenow/3210/02.html Mechanism of RNAi in mammalian cells Antonin et. al ., 2007 Nature Reviews Drug Discovery 6, 443-453 Rational siRNA design 5’-P 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 A U xG A 3’’- OH P- 5’’ 3’’- OH High thermal stability of the 5’ sense strand (SS) blocks incorporation of SS into RISC Low thermal stability of the 5’ anti-sense strand (AS) promotes Incorporation of AS into RISC. G or C is preferred. AU rich is suggested. Low stability in this region enhances RISC/AS-mediated cleavage of mRNA and promote RISC complex release. U at position 10 at SS is recommended. Sense strand Antisense strand Reynolds et al. 2004 Other considerations no high GC content (35-65%); no inverted repeat sequence; no consecutive 3 Gs or 3 Cs; no consecutive 4 Ts if use polIII promoter; Rational siRNA design Crite ria Description Score 1 Moderate to low (30%-52%) GC Content Yes 1 point 2 At least 3 A/Us at positions 15-19 (sense) 3 Lack of internal repeats (Tm*<20¡ãC) 1 point /per A or U 1 point 4 A at position 19 (sense) 1 point 5 A at position 3 (sense) 1 point 6 U at position 10 (sense) 1 point 7 No G/C at position 19 (sense) -1 point 8 No G at position 13 (sense) -1 point No These characteristics are used by rational siRNA design algorithm to evaluate potential targeted sequences and assign scores to them. Sequences with higher scores will have higher chance of success in RNAi. Reynolds et al. 2004 Phases of TRC Program The RNAi Consortium (TRC) The RNAi Core Phase I (May/2004 to Apr/2007) Jun/2005 to Apr/2008 Phase II (Oct/2007 to Sept/2011) May/2008 to Apr/2011 Vector Used by RNAi Core http://www.sigmaaldrich.com/Area_of_Interest/Life_Science/Functional_Genomics_and_RNAi/Product_Lines/shRNA_Library.html Name Description U6 Promoter RNA generated with four uridine overhangs at each 3' end Cppt/CTE Central polypurine tract /constitutive transport element hPGK Human phosphoglycerate kinase eukaryotic promoter puroR Puromycin resistance gene for mammalian selection SIN/LTR 3' self inactivating long terminal repeat f1 ori f1 origin of replication ampR Ampicillin resistance gene for bacterial selection pUC ori 5' LTR Psi RRE pUC origin of replication 5' long terminal repeat RNA packaging signal Rev response element Configuration of TRC shRNA construct Materials Received from TRC shRNA constructs and knockdown information: TRC-I TRC-II Clone # Gene # Clone # Gene # Knockdown Information Human 83,117 16,026 53,070 10,149 41,774#1 Mouse 79,200 15,976 44,042 9,027 34,325#2 162,317 32,002 97,112 19,176 76,099 Total #1 Targeting 7,074 genes #2 Targeting 6,738 genes 35 shRNA expression vectors (some are intermediates). • with different selection/ fluorescence markers 04-11-2010 updated TRC library performance Good hairpins/gene coverage Highest quality data: 10,711 genes 25% 16000 14000 20% % genes 10000 8000 15% 10% 6000 5% 4000 2000 100-90 90-80 80-70 %KD 70-50 No KD ~0/5 ~1/5 ~2/5 ~3/5 0 ~4/5 0% all HPS #shRNA 12000 "good" clones/gene TRC report TRC validation (QRT-PCR) Tests on 30,000 hairpin panels (~150,000 tests) Released data on 14K distinct genes 35,000 Process upgrade successful attempted 25,000 1,200 Genes / month (6,000 shRNAs) 20,000 15,000 10,000 5,000 0 September February April May June July August September October November December January February March April May June July August September October November December January February March April May June July August September October November December January # genes (cumulative) 30,000 Resume 2006 2007 2008 2009 Throughput: 1,200 2,000 genes/month 2010 TRC report Available Lentivirus Item (arrayed, pooled, individual) human kinase and phosphatase (hKP) subset Mouse kinase subset / Mouse phosphatase subset Human/ Mouse tumor suppressor subset Human / Mouse transcription Factor subset Human deubiqutinating enzymes subset Control viruses Pooled hKP Pooled mouse kinase / Pooled mouse phosphatase Pooled Human/ Mouse tumor suppressor set Pooled Human / Mouse transcription Factor subset Pooled Human deubiqutinating enzymes subset Customized VSV-G pseudotyped lentivirus 80K Human shRNA Pool (16,026 genes) Migration Profile of hTRC1 Pooled pDNA Pool # Ctl M 1 2 3 4 5 6 7 8 9 10 M 10K 6K 4K 3K 2K 86723 Clones 17745 Genes 80264 TRC1 Human Clones 16026 TRC1 Human Genes 4713 TRC1 Mouse Clones 1377 TRC1 Mouse Genes 1661 TRC2 New Human Clones 342 TRC2 New Human Genes 85 Control Clones Expression of Hairpin RNA (shRNA) Using Pol III Promoters Transcription initiation of DNA-dependent RNApol III promoters (U6 or H1) are well characterized. RNApol III transcription uses a well-defined termination signal (TTTTT) and the products have no extra sequence. Transcription from these promoters is very efficient in various tissues. Configuration/Structure of hU6 Promoter -23 TATATAT DSE PSE ≈ 250bp +1 -48 Structure of VSV-G-Pseudotyped Lentivirus Modified from http://www.washington.edu/alumni/columns/dec00/cells4.html Replication of Retrovirus http://www.accessexcellence.org/RC/VL/GG/retrovirus.html Genome Organization of Lentiviral Vector (Improved biosafety by eliminating non-essential genes or sequences) pLKO.1-puro: RSV promoter Psi signal R-U5 RRE pbs pCMVΔR8.91: SD Gag Pol hPGK promoter AgeI EcoRI U3-R U6 promoter Puro SA SD SA Rev ΔΨ VSV-G pMD.G: CMV promoter SV40 PolyA RRE Tat CMV promoter PPT SIN SV40 PolyA SV40 PolyA HEK293T as Packaging Cells Procedures: Day1: seeding cells Day2: co-transfection Day3: re-fresh media Day4: harvest viruses/ re-add media Day5: harvest viruses From genome sequence to gene function Function Genomics What does the gene mean? Forward and reverse genetics approach to study gene function Forward Genetics: start with a phenotype, find the gene. naturally occurring mutants can be used. Reverse Genetics: start with a gene, determine its phenotype. identify the phenotypes resulting from the disruption of a particular gene. Applications of RNAi libraries RNA Interference Cancer • • • • • Basic Research Viral Infection Gene functions Tumor biology Host factors required for viral replication Biological pathways And more Approaches to large-scale RNAi screen/selection Arrayed RNAi library/Screen siRNA Plasmid Vector Viral Vector Pooled RNAi library /Selection Viral Vector Transfection Transduction High Throughput Assay for Altered Phenotype(s) 2nd assay to validate hits Transduction Selective screen for Altered Phenotype(s) Hits identification: Barcode microarray/ RT-PCR sequencing RNAi pooled screening: positive selection R&D in RNAi SAEC/Core Control Search for cellular factors that support primary human small airway epithelial cell (SAEC) growth using RNAi pooled selection, 17 genes that support SAEC growing in soft agar are identified. SAEC/ Control SAEC/ RNAi > 0.45 mm SAEC/ RNAiAnchorage-dependent growth assay RNAi pooled screening: negative selection How to ensure that hits aren't off-target ◙ Off-Target: ◙ How/ Criterion: Phenotype change is caused by two or more independent shRNAs that target the same gene ◙ Why: Degradation of mRNA can occur by two separate pathways in RNAi Khvorova A. RNA (2008),14:853-861. 3’ UTR hexamer frequency in human genome SCF: seed complementary frequency high(>3800), medium (z2500–2800), or low (<350) SCFs in the HeLa transcriptome Khvorova A. RNA (2008),14:853-861. Microarray signatures of GAPDH- and PPIB-targeting siRNAs One nt shift in seed sequence: GAPDH M1 sense: 5GGCUCACAACGG GAAGCUU GAPDH M8 sense: 5GCUCACAACGGG AAGCUUG Seed region not static Same seed sequences in different target genes: GAPDH H15 sense: 5-GAAGUAUGACAACAGCCUC PPIB H17 sense: 5-CGACAGUCAAGACAGCCUG Khvorova A. RNA (2008),14:853-861. Seed sequence plays major role in off-target Khvorova A. RNA (2008),14:853-861. Configuration of TRC shRNA construct shRNA processing How are the TRC library shRNAs processed into short dsRNAs? Implications: hairpin design, off-target effects polIII transcription start and stop; evidence for DROSHA processing? GGGTCGAGCTGGACGGCGACGTAC TTTTTCAGCTCGACCTGCCGCTGCATG Where does DICER cut? T A C G 22 nts Which strand goes into RISC? (Strand that goes into RISC is more stable/abundant) TRC: Jen Grenier, Andrew Grimson, Ozan Alkan Small RNA sequencing: all 26 shRNAs Length#reads % shRNA % strand GG GG GG GG 23mer 5,217 22mer 32,279 21mer 8,029 20mer 1,029 G21merAntisenseStrandSTTTTT (4) (3) A (5) T 11% 67% 17% 2% C G GG21merSenseStrandSeqncC 1% 7% 2% <1% 22mer 21mer 20mer 23mer 22mer 21mer 23mer 18,285 4% 39,095 9% 6,760 2% 45,610 10% 205,249 46% 40,444 9% 23,263 5% 5% 10% } 17% 2% 11% 51% } 72% 10% 6% e e e m r r r 3Ts 4Ts 5Ts 3Ts 4Ts 5Ts 4Ts Highly parallel identification of essential genes in cancer cells Biao Luo etc, Proc Natl Acad Sci U S A, 2008, 105: 20380–20385. Pooled RNAi screening (45K lentiviruses) Pooled RNAi screening (45K lentiviruses) Involed in FAS induced apoptosis Screens for essential genes in 12 cancer cell lines Commonly essential gene NSCLC glioblastoma SCLC leukemia Cell lineage-specific essential gene Cell line-specific essential gene (Cancer specific gene dependency) Time course analysis for the top 100 essential genes in K562 cells Integration of functional and structural genomics (Case in NSCLC) Integration of functional and structural genomics (Case in NSCLC) Integration of functional and structural genomics (Case in NSCLC) References: Review paper & original paper 1. Oncogene (2004) 23:8346-8352; 8376-8383; 8384-8391; 8392-8400; 8401-8409. 2. Moffat J & Sabatini DM 2006. Building mammalian signaling pathways with RNAi screens. Nature Rev. 7:177-187. 3. Focus on RNA interference (a user guide). Nature Methods 2006 Sep 3(9):669-719. 4. Paddison PJ (2008) RNA interference in mammalian cell systems. Curr Top Microbiol Immunol. 2008;320:1-19. 5. Recent reviews on RNAi. Curr Top Microbiol Immunol 2008, volume 320:1-201. 6. Luo Biao et al. 2008. Highly parallel identification of essential genes in cancer cells. Proc Natl Acad Sci U S A. 105(51): 20380–20385. National RNAi Core website http://rnai.genmed.sinica.edu.tw/