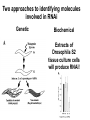
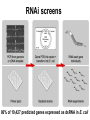
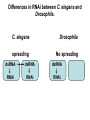
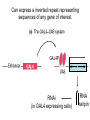
* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download RNAi
Deoxyribozyme wikipedia , lookup
Molecular evolution wikipedia , lookup
Messenger RNA wikipedia , lookup
Genome evolution wikipedia , lookup
Community fingerprinting wikipedia , lookup
List of types of proteins wikipedia , lookup
Polyadenylation wikipedia , lookup
RNA polymerase II holoenzyme wikipedia , lookup
X-inactivation wikipedia , lookup
Eukaryotic transcription wikipedia , lookup
Gene desert wikipedia , lookup
Gene therapy wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Gene expression profiling wikipedia , lookup
Endogenous retrovirus wikipedia , lookup
Promoter (genetics) wikipedia , lookup
Transcriptional regulation wikipedia , lookup
Gene regulatory network wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Non-coding RNA wikipedia , lookup
Epitranscriptome wikipedia , lookup
Gene expression wikipedia , lookup
Silencer (genetics) wikipedia , lookup