* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Figure 1 - genomics-lab
DNA barcoding wikipedia , lookup
Holliday junction wikipedia , lookup
Human genome wikipedia , lookup
Cancer epigenetics wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Genetic drift wikipedia , lookup
Frameshift mutation wikipedia , lookup
Zinc finger nuclease wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
DNA sequencing wikipedia , lookup
DNA profiling wikipedia , lookup
DNA vaccination wikipedia , lookup
DNA damage theory of aging wikipedia , lookup
Comparative genomic hybridization wikipedia , lookup
United Kingdom National DNA Database wikipedia , lookup
History of genetic engineering wikipedia , lookup
Genomic library wikipedia , lookup
Primary transcript wikipedia , lookup
Dominance (genetics) wikipedia , lookup
Molecular cloning wikipedia , lookup
Metagenomics wikipedia , lookup
Extrachromosomal DNA wikipedia , lookup
Non-coding DNA wikipedia , lookup
DNA polymerase wikipedia , lookup
Nucleic acid double helix wikipedia , lookup
DNA supercoil wikipedia , lookup
Epigenomics wikipedia , lookup
Genealogical DNA test wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Microevolution wikipedia , lookup
Point mutation wikipedia , lookup
Helitron (biology) wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Gel electrophoresis of nucleic acids wikipedia , lookup
Molecular Inversion Probe wikipedia , lookup
Cell-free fetal DNA wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Bisulfite sequencing wikipedia , lookup
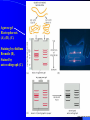
f. Microsatellites or SSR Markers In 1989, Weber & May and Litt & Luty discover microsatellite sequences, demonstrate their high level of polymorphism due to variations in the number of tandem repeats (1 - typical heterozygosities in cattle), abundance and even distribution across the genome. Microsatellites are genotyped using the polymerase chain reaction (1 ) using primers targeted to the unique sequences flanking the microsatellite motif. PCR can easily be semi-automated (1 ) The resulting PCR products are separated according to size by gel electrophoresis using either agarose gels or more commonly (because of their higher resolution) denaturing polyacrylamide gels (PAGE) (1 ). PCR products are visualized by: Fig. Tri-nucleotide repeat microsatellite marker: Expansion of the CGG triplet in the FMR-1 gene seen in the fragile X syndrome. Normal individuals have from 6 to 54 copies of the CGG repeat, while individuals from susceptible families display an increase (premutation) in the number of repeats: normally transmitting males (NTMs) and their daughters are phenotypically normal but display 50 to 200 copies of the CGG triplet; the number of repeats expands to some 200 to 1300 in individuals showing full symptoms of the disease. PCR for semi-automated Microsatellite genotyping by auto-radiography. Denaturated Poly-acrylamide gel electrophoresis (PAGE) for microsatellite genotyping •Direct staining (ethidium bromide or silverstaining)(1 ) •Autoradiography: •The PCR products are labelled either by incorporation of [a -P 32 or 33] dNTPs (1 )or [a -S 35] dNTS during the PCR amplification (1 )(labels the two strands), or by using one end-labeled primer (labels one strand). Primers are endlabeled using [g -P 32 ]ATP & T4 polynucleotide kinase (1 ) •After gel electrophoresis, an X-ray film is exposed to the gel revealing the position of the PCR products as black spots. A photon of light or a b particle or g ray released from a radioactive molecule "activate" silver bromide crystals on the film emulsion. This renders them capable of being reduced through the developing process to form silver metal (a "grain"). The silver grains on the film form the image. •Co-amplification and/or co-loading of multiples microsatellites allows for multiplex genotyping (2-4 systems). Agarose gel Electrophoresis (A), (B), (C). Staining by ethidium Bromide (B), Stained by auto-radiograph (C). Labeling of genetic markers Labelling of dNTP by P32 radio-isotops in microsatellite tying by auto-radiogragy. Mode of action of P32 labeling in PCR product. •Fluorescence labelling: •The PCR products are labeled either by using primers or dNTPs which are tagged with an appropriate fluorophore, a chemical group which fluoresces when exposed to a specific wavelength of light. Popular fluorophores used in direct labeling include fluorescein, a pale green fluorescent dye, rhodamine, a red fluorescent dye, and amino methyl coumarin, a blue fluorescent dye. Fluorophoes are characterized by their excitation and emission spectra. •The PCR products are detected during migration using automatic sequencers. (1 ) •Co-amplification and/or co-loading of multiples microsatellites allows for multiplex genotyping (up to 20 systems). (1 ) •Software packages allow for semi-automated data capture (1 ). Microsatellite profiles are often difficult to read due to artefactual bands, which result from (1 ): •the differential migration of the two DNA strands in denaturing acrylamide gels •the "+ A" activity of the Taq polymerase generating x + 1 bands •slippage of the Taq polymerase during polymerization generating … +4, +2, -2, -4, … "stutter" bands •non denatured secondary structures adopted by the PCR products. Microsatellite typing by automated DNA sequencer: New era of molecular genetics: Using fluorescent labeled primers.(4Pictures of ABI377 PerkinElmer DNA sequencers) g.SNPs (Single nucleotide polymorphisms) The difficulty to fully automate microsatellite genotyping has revived interest in a new type of markers: single nucleotide polymorphisms or SNPs. Definition: SNPs are polymorphisms due to single nucleotide substitutions (transitions > transversions) or single nucleotide insertions/deletions. Abundance: The average heterozygosity per nucleotide site, p , has been estimated at approximately 1/1000 in man, 1/2500 in cattle. Informativeness: SNPs are virtually always biallelic markers. Their heterozygosity is therefore limited at 50%. Examples of SNP genotyping methods: •1) Single Stranded Conformation Polymorphism (SSCP)(1 ) •2) Allele specific oligonucleotides (ASO)(1 ) •3) Single nucleotide polymorphic discrimination by an electronic dot blot assay (ASO) on semconductor microchips (1 ; 1 ) •4) Reverse dot blot on DNA chips (1 ) •5) Dynamic allele specific hybridisation (DASH) (1 ; 1 ) •6) Allele-specific PCR (=amplification refractory mutation system or ARMS test)(1 ) •7) Mutation detection the ARMS test in combination with the TaqmanTM 5' exonuclease assay (exploiting the 5'->3' exonuclease activity of Taq DNA polymerase).(1 ) •8) Minisequencing and analysis of the extension products by PAGE. •9) Minisequencing and analysis of the extension products on DNA chips •10) Minisequencing and analysis of the extension products using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDITOF)(1 ) •11) Pyrosequencing (1 ) •12) OLA (1 ) •13) Invasive clivage of oligonucleotide probes (Invader technology) •1) Single Stranded Conformation Polymorphism (SSCP)(1 (Figure 1-3) Detection and production of Allele specific Oligo-nucleotide (ASO) Allele-specific oligonucleotide (ASO) dot-blot hybridisation can identify individuals with the sickle cell mutation. The sickle cell mutation is a single nucleotide substitution (A to T) at codon 6 in the b -globin gene, resulting in a GAG (Glu) to GTG (Val) substitution. The example shows how one can design ASOs: one specific for the normal (b A) allele and identical to a sequence of 19 nucleotides encompassing codons 3-9 of this allele, and one specific for the mutant (b S) allele, being identical to the equivalent sequence of the mutant allele. The labeled ASOs can be individually hybridized to denatured genomic DNA samples on dot-blots. The b A- and b S-specific ASOs can hybridize to the complementary antisense strand of the normal and mutatnt alleles respectively, forming perfect 19-bp duplexes. However, duplexes between the b A-specific ASO and the b S allele, or between the b S-specific ASO and the b A allele have a single mismatch and are unstable at high hybridization stringency. •2) Single nucleotide polymorphic discrimination by an electronic dot blot assay (ASO) on semiconductor microchips •Single nucleotide polymorphic discrimination by an electronic dot blot assay (ASO) on semiconductor microchips Dynamic allele-specific hybridization (DASH) Figure 1: DASH assay principle. Probe-target denaturation is monitored over a wide temperature range by measuring fluorescence while heating. All tested sequences thus yield a melting temperature indicative of either perfect match or single-base mismatch to the assay probe. Dynamic allele-specific hybridization W. Mathias Howell et al. Nature Biotechnology 17, 87 - 88 (1999) Correct base-pairing at the 3' end of PCR primers is the basis of allele-specific PCR. The allele-specific oligonucleotide primers ASP1 and ASP2 are designed to be identical to the sequence of the two alleles over a region preceding the position of the variant nucleotide, up to and terminating in the variant nucleotide itself. ASP1 will bind perfectly to the complementary strand of the allele 1 sequence permitting amplification with conserved primer. However, the 3'-terminal C of the ASP2 primer mismatches with the T of the allele 1 sequence, making amplification impossible. Similarly ASP2 can bind perfectly to allele 2 and initiate amplification, unlike ASP1. The TaqmanTM 5' exonuclease assay (Mutation detection by ARMS) In addition to two conventional PCR primers, P1 and P2, which are specific for the target sequence (P1 being for instance allele specific), a third primer, P3 is designed to bind specifically to a site on the target sequence downstream of the P1 binding. P3 is labeled with two fluorophores, a reporter dye (R) is attached at the 5' end, and a quencher dye (D), which has a different emission wavelength to the reporter dye, is attached at its 3' end. Because its 3' end is blocked, primer P3 cannot by itself prime any new DNA synthesis. During the PCR reaction, Taq DNA polymerase synthesizes a new DNA strand primed by P1 and as the enzyme approaches P3, its 5'-> 3' exonuclease activity processively degrades the P3 primer from its 5' end. The end result is that the nascent DNA strand extends beyond the P3 binding site and the reporter and quencher dyes are no longer bound to the same molecule. As the reporter dye is no longer in close proximity to the quencher, the resulting increase in reporter emission intensity is easily detected. Figure 1: Mass compression for a sixfold multiplex genotyping assay. Loci 2, 3, 5, 7, 8, and 10 were amplified and typed using corresponding primers and standard assay conditions. Arrows indicate corresponding primer-extended primer pairs. Molecular weight assignments from two point calibration are given. High level multiplex genotyping by MALDI-TOF mass spectrometry Philip Ross et al. Nature Biotechnology 16, 1347 - 1351 (1998) Pyrosequencing™ is sequencing by synthesis. ' A simple to use technique for accurate and consistent analysis of large numbers of short to medium length DNA sequences. Step1. A sequencing primer is hybridized to a single stranded, PCR amplified, DNA template, and incubated with the enzymes, DNA polymerase, ATP sulfurylase, luciferase and apyrase, and the substrates, adenosine 5´ phosphosulfate (APS) and luciferin. Polymorphism identification and quantitative detection of genomic DNA by invasive cleavage of oligonucleotide probes Victor Lyamichev et al. Nature Biotechnology 17, 292 - 296 (1999) Figure 1: Schematic representation of the configurations assumed by oligonucleotides with an overlap complementary to a DNA target. The 3´ end of the upstream oligonucleotide (A) or the 5´ end of the downstream oligonucleotide (C) can each be displaced by the other, with both conformers interchanging through an intermediate form (B). The FEN1 endonucleases used in this study cleave only the flap created by displacement of the downstream segment (C).