* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download 14.1 The lacI Gene Encodes a Diffusible Repressor
Cancer epigenetics wikipedia , lookup
Oncogenomics wikipedia , lookup
Genomic imprinting wikipedia , lookup
Pathogenomics wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Protein moonlighting wikipedia , lookup
Gene desert wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Frameshift mutation wikipedia , lookup
Gene therapy wikipedia , lookup
Neuronal ceroid lipofuscinosis wikipedia , lookup
Genetic engineering wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Genome evolution wikipedia , lookup
Saethre–Chotzen syndrome wikipedia , lookup
Gene nomenclature wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Epigenetics of human development wikipedia , lookup
History of genetic engineering wikipedia , lookup
Helitron (biology) wikipedia , lookup
X-inactivation wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Genome (book) wikipedia , lookup
Gene expression profiling wikipedia , lookup
Gene expression programming wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Designer baby wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Microevolution wikipedia , lookup
Point mutation wikipedia , lookup
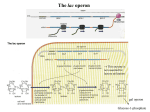
Robert J. Brooker - Genetica Esperimento di genetica 14.1 The lacI Gene Encodes a Diffusible Repressor Protein Now that we have an understanding of the lac operon, let’s consider one of the experimental approaches that was used to elucidate its regulation. In the 1950s, Jacob, Monod, and their colleague Arthur Pardee had identified a few rare mutant strains of bacteria that had abnormal lactose adaptation. One type of mutant, designated lacI–, resulted in the constitutive expression of the lac operon even in the absence of lactose. From this observation, the researchers incorrectly hypothesized that the lacI– mutation resulted in the synthesis of an internal inducer, making it unnecessary for cells to be exposed to lactose for induction (Figure EG14.1.1). By comparison, Figure EG14.1.1b shows the correct explanation. A loss-of-function mutation in the lacI gene prevented the lac repressor protein from inhibiting transcription. At the time of their work, however, the function of the lac repressor was not yet known. To further explore the nature of this mutation, Jacob, Monod, and Pardee applied a genetic approach. In order to understand their approach, let’s briefly consider the process of bacterial conjugation (described in Chapter 6). The earliest studies of Jacob, Monod, and Pardee in 1959 involved matings between recipient cells, termed F–, and donor cells, which were Hfr strains that transferred a portion of the bacterial chromosome. Later experiments in 1961 involved the transfer of circular segments of DNA known as F factors. We will consider the latter type of experiment here. Sometimes an F factor also carries genes that were originally found within the bacterial chromosome. These types of F factors are called F' factors (F prime factors). In their studies, Jacob, Monod, and Pardee identified F' factors that carried the lacI gene and portions of the lac operon. These F' factors can be transferred from one cell to another by bacterial conjugation. A strain of bacteria containing F' factor genes is called a merozygote, or partial diploid. The production of merozygotes was instrumental in allowing Jacob, Monod, and Pardee to elucidate the function of the lacI gene. This experimental approach has two key points. First, the two lacI genes in a merozygote may be different alleles. For example, the lacI gene on the chromosome may be a lacI– allele that causes constitutive expression, while the lacI gene on the F' factor may be normal. Second, the genes on the F' factor and the genes on the bacterial chromosome are not physically adjacent to each other. As we now know, the expression of the lacI gene on an F' factor should produce repressor proteins that could diffuse within the cell and eventually bind to the operator site of the lac operon located on the chromosome. Figure EG14.1.2 shows one experiment of Jacob, Monod, and Pardee in which they analyzed a lacI– mutant strain that was already known to constitutively express the lac operon and compared it to the corresponding merozygote. The merozygote had a lacI– mutant gene on the chromosome and a normal lacI gene on an F' factor. These two strains were grown and then divided into two tubes each. In half of the tubes, lactose was omitted. In the other tubes, the strains were incubated with lactose to determine if lactose was needed to induce the expression of the operon. The cells were lysed by sonication, and then a lactose analogue, β-ONPG, was added. This molecule is colorless, but β-galactosidase cleaves it into a product that has a yellow color. Therefore, the amount of yellow color produced in a given amount of time is a measure of the amount of β-galactosidase that is being expressed from the lac operon. THE HYPOTHESIS The lacI– mutation results in the synthesis of an internal inducer. FIGURE EG14.1.1 Alternative ideas to explain how a lacI– mutation could cause the constitutive expression of the lac operon. (a) The hypothesis of Jacob, Monod, and Pardee. In this case, the lacI– mutation would result in the synthesis of an internal inducer that turns on the lac operon. (b) The correct explanation in which the lacI– mutation eliminates the function of the lac repressor protein, which prevents it from repressing the lac operon. © 2010 The McGraw-Hill Companies, S.r.l. - Publishing Group Italia Robert J. Brooker - Genetica TESTING THE HYPOTHESIS — FIGURE EG14.1.2 Evidence that the lacI gene encodes a diffusible repressor protein. Starting material: The genotype of the mutant strain was lacI– lacZ+ lacY+ lacA+. The merozygote strain had an F' factor that was lacI+ lacZ+ lacY+ lacA+, which had been introduced into the mutant strain via conjugation. © 2010 The McGraw-Hill Companies, S.r.l. - Publishing Group Italia Robert J. Brooker - Genetica THE DATA Strain Mutant Mutant Merozygote Merozygote Addition of Lactose No Yes No Yes Amount of β-Galactosidase (percentage of parent strain) 100% 100% <1% 220% INTERPRETING THE DATA As seen in the data, the yellow production in the original mutant strain was the same in the presence or absence of lactose. This result is expected because the expression of β-galactosidase in the lacI– mutant strain was already known to be constitutive. In other words, the presence of lactose was not needed to induce the operon due to a defective lacI gene. In the merozygote strain, however, a different result was obtained. In the absence of lactose, the lac operons were repressed—even the operon on the bacterial chromosome. How do we explain these results? Because the normal lacI gene on the F' factor was not physically located next to the chromosomal lac operon, this result is consistent with the idea that the lacI gene codes for a repressor protein that can diffuse throughout the cell and bind to any lac operon. The hypothesis that the lacI– mutation resulted in the synthesis of an internal inducer was rejected. If that hypothesis had been correct, the merozygote strain would have still made an internal inducer, and the lac operons in the merozygote would have been expressed in the absence of lactose. This result was not obtained. The interactions between regulatory proteins and DNA sequences illustrated in this experiment have led to the definition of © 2010 The McGraw-Hill Companies, S.r.l. - Publishing Group Italia Robert J. Brooker - Genetica two genetic terms. A trans-effect is a form of genetic regulation that can occur even though two DNA segments are not physically adjacent. The action of the lac repressor on the lac operon is a trans-effect. A regulatory protein, such as the lac repressor, is called a trans-acting factor. In contrast, a cis-acting element is a DNA segment that must be adjacent to the gene(s) that it regulates, and it is said to have a cis-effect on gene expression. The lac operator site is an example of a cis-acting element. A trans-effect is mediated by genes that encode regulatory proteins, whereas a cis-effect is mediated by DNA sequences that are bound by regulatory proteins. Jacob and Monod also isolated constitutive mutants that affected the operator site, lacO. Table EG14.1.1 summarizes the effects of mutations based on their locations in the lacI regulatory gene versus lacO and their analysis in merozygotes. As seen here, a loss-of-function mutation in a gene encoding a repressor protein has the same effect as a mutation in an operator site that cannot bind a repressor protein. In both cases, the genes of the lac operon are constitutively expressed. In a merozygote, however, the results are quite different. When a normal lacI gene and a normal lac operon are introduced into a cell harboring a defective lacI gene, the normal lacI gene can regulate both operons. In contrast, when a lac operon with a normal operator site is introduced into a cell with a defective operator site, the operon with the defective operator site continues to be expressed without lactose present. Overall, a mutation in a trans-acting factor can be complemented by the introduction of a second gene with normal function. However, a mutation in a cis-acting element is not affected by the introduction of another cis-acting element with normal function into the cell. TABLE EG14.1.1 A Comparison of Loss-of-Function Mutations in the lacI Gene versus the Operator Site Chromosome Wild type lacI– lacO– lacI– lacO– F’ factor None None None lacI+ and a normal lac operon lacI+ and a normal lac operon © 2010 The McGraw-Hill Companies, S.r.l. - Publishing Group Italia Expression of the lac Operon With Lactose Without Lactose 100% <1% 100% 100% 100% 100% 200% <1% 200% 100%