* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download RNA Transcription
Alternative splicing wikipedia , lookup
Bottromycin wikipedia , lookup
Histone acetylation and deacetylation wikipedia , lookup
Protein adsorption wikipedia , lookup
List of types of proteins wikipedia , lookup
Protein (nutrient) wikipedia , lookup
Gene regulatory network wikipedia , lookup
RNA interference wikipedia , lookup
Non-coding DNA wikipedia , lookup
Transcription factor wikipedia , lookup
Molecular evolution wikipedia , lookup
RNA silencing wikipedia , lookup
Biochemistry wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Point mutation wikipedia , lookup
Polyadenylation wikipedia , lookup
Promoter (genetics) wikipedia , lookup
Expanded genetic code wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Non-coding RNA wikipedia , lookup
Genetic code wikipedia , lookup
Messenger RNA wikipedia , lookup
Gene expression wikipedia , lookup
Eukaryotic transcription wikipedia , lookup
RNA polymerase II holoenzyme wikipedia , lookup
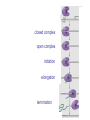
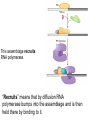
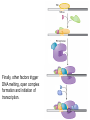
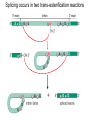
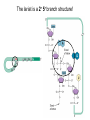
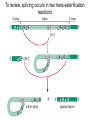
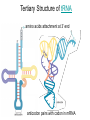
October 9 Richard Losick Where we’ve been and where we are going! • DNA structure and DNA replication Today and Next Tuesday • Gene transcription and the translation of RNA into protein Final two lectures • How genes are switched ON and OFF • How life arose on Earth, if it did arise on Earth! Gene transcription and mRNA maturation 1. Central dogma and gene transcription 3. RNA polymerase 4. mRNA maturation in eukaryotes Goal: To understand how genetic information is transmitted from the genome to the ribosome. Objectives: You should be able to: • compare and contrast transcription with replication. • describe the transcription machinery. • describe how transcripts are processed into mRNAs. 2. Central Dogma and Gene Transcription "The central dogma states that once 'information' has passed into protein it cannot get out again. The transfer of information from nucleic acid to nucleic acid, or from nucleic acid to protein, may be possible, but transfer from protein to protein, or from protein to nucleic acid, is impossible. Information means here the precise determination of sequence, either of bases in the nucleic acid or of amino acid residues in the protein.“ Francis Crick, 1958 The Central Dogma states that information flows from nucleic acids into protein Transcription takes place asymmetrically in a moving bubble (There is no “lagging” strand) Non-template strand ? Template strand Is the question mark the 3’ or 5’ terminus? 3. RNA polymerase RNA polymerase recognizes punctuation marks in the DNA known as promoters RNA polymerase in bacteria recognizes sequences located 10 (“-10”) and 35 (“-35”) base pairs upstream of the start site of transcription (+1) TTGACA and TATAAT are a Platonic ideal! closed complex open complex in which 14 bp of DNA are unwound Promoter recognition is built into bacterial RNA polymerase. initiation when the first phosphodiester bond is formed elongation termination closed complex open complex with ~13 base pairs unwound initiation when the first phosphodiester bond is formed elongation termination closed complex open complex initiation – formation of the first phosphodiester bond elongation termination closed complex open complex initiation elongation termination closed complex open complex initiation elongation termination Crab claw demonstration Promoter recognition works differently in eukaryotes! Eukaryotic RNA polymerase does not recognize promoters. Instead, “transcription factors” recruit RNA polymerase. The most important is TATA- binding protein (TBP). TBP recognizes a sequence called the TATA box. TATA-binding protein binds in the minor groove and causes the DNA to bend. TATA-binding protein (TBP) binds to the TATA box. Next, TBP recruits additional transcription factors to the DNA (colored shapes). This assemblage recruits RNA polymerase. “Recruits” means that by diffusion RNA polymerase bumps into the assemblage and is then held there by binding to it. Finally, other factors trigger DNA melting, open complex formation and initiation of transcription. Main points 1. Transcription is asymmetric and occurs with the release of the RNA transcript. 2. Transcription is catalyzed by RNA polymerase, which binds to promoters. 3. Bacterial promoters consist of -10 and -35 sequences. 4. In eukaryotes, transcription factors, such as the TATA-binding protein, bind to the promoter and recruit RNA polymerase. 4. mRNA maturation in eukaryotes mRNA undergoes maturation events before it exits the nucleus and is translated: CAPing Tailing Splicing Transcripts acquire a “CAP” at their 5’ end. The CAP is a guanine nucleotide joined to mRNA by a 5’-5’ linkage. 3’-G-5’ppp5’-N-p........ Transcripts acquire a tail of about 200 As at their 3’ terminus. exon CAP intron exon (A) n Coding sequences (“exons”) in mRNA are interrupted by non-coding sequences, “introns”, which are removed before the mRNA is translated. • Eukaryotic genes are in pieces. • The coding segments are exons and the interruptions are introns. • The introns are removed by “splicing” after the gene is transcribed into RNA. Splicing occurs in two trans-esterification reactions First, the 2’ hydroxyl of the branch point A attacks the phosphoryl group of the G at the 5’ splice site to create a lariat. The lariat is a 2’ 5’ branch structure! The 3’ OH of the 5’ exon becomes the nucleophile in the 2nd trans-esterification reaction. This joins the 5’ to the 3’ exon, releasing of the lariat. To review, splicing occurs in two trans-esterification reactions Story time! Joan Steitz Translation Goal: To understand how mRNA is translated into protein. Objectives: You should be able to: • describe the principal features of the genetic code. • describe tRNA charging and how fidelity is achieved. • describe the principal features of the ribosome. • explain the translation cycle and how fidelity is achieved. • explain how open-reading frames are set The genetic code translates the 4-letter alphabet of mRNA into the 20-word language of protein. What is the minimum number of nucleotides needed to specify 20 amino acids? Is it two, three, four…? If mRNA were read in units of two nucleotides, it could specify only 16 (42) amino acids –too few! Ergo, messenger RNA must be read in units of (at least) three nucleotides. If it were read in units of the three, the number of permutations would be 43 or 64. The Rosetta Stone of Life! Key features of the genetic code Codons are three nucleotides long. Codons are read by the ribosome in a 5’ to 3’ direction: 5’-XXX-3’. 61 triplets specify amino acids. The remaining three are stop codons. The Genetic Code How is the 4-base language of mRNA translated into the 20-amino acid language of protein? tRNAs are adaptors between codons and amino acids. Each tRNA is “charged” by a specific amino acid and recognizes a particular codon. Cloverleaf Structure of tRNA amino acid attachment at 3’ end anticodon pairs with codon in mRNA Tertiary Structure of tRNA amino acids attachment at 3’ end anticodon pairs with codon in mRNA