* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Association Studies and High-throughput Genotyping Technologies
Gene desert wikipedia , lookup
X-inactivation wikipedia , lookup
Metagenomics wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Genetic testing wikipedia , lookup
Polymorphism (biology) wikipedia , lookup
Molecular Inversion Probe wikipedia , lookup
Genomic library wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Gene expression programming wikipedia , lookup
Medical genetics wikipedia , lookup
Minimal genome wikipedia , lookup
Copy-number variation wikipedia , lookup
Whole genome sequencing wikipedia , lookup
Genomic imprinting wikipedia , lookup
Genetic engineering wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Hardy–Weinberg principle wikipedia , lookup
Human genome wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Pathogenomics wikipedia , lookup
History of genetic engineering wikipedia , lookup
Genome editing wikipedia , lookup
Designer baby wikipedia , lookup
Quantitative trait locus wikipedia , lookup
Behavioural genetics wikipedia , lookup
Heritability of IQ wikipedia , lookup
Genetic drift wikipedia , lookup
Dominance (genetics) wikipedia , lookup
Genome evolution wikipedia , lookup
Population genetics wikipedia , lookup
SNP genotyping wikipedia , lookup
Human genetic variation wikipedia , lookup
Genome (book) wikipedia , lookup
Pharmacogenomics wikipedia , lookup
A30-Cw5-B18-DR3-DQ2 (HLA Haplotype) wikipedia , lookup
Microevolution wikipedia , lookup
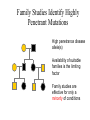
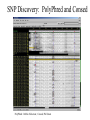
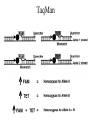
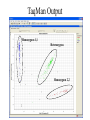
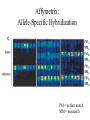
MEDG 505 Pharmacogenomics March 17, 2005 A. Brooks-Wilson Reminder: What is Genomics? According to http://genomics.ucdavis.edu/what.html: “Genomics is operationally defined as investigations into the structure and function of very large numbers of genes undertaken in a simultaneous fashion” Pharmacogenetics • “The study of how genes affect people’s response to medicines” (NIH) • A subset of complex genetics for which the traits relate to drugs • First observed in 1957 • Part of “personalized medicine” • 20-95% of variability in drug disposition and effects is thought to be genetic • Non-genetic factors: age, interacting medications, organ function • Drug absorption, distribution, metabolism, excretion • >30 families of genes Pharmacogenetics: Examples • Drug metabolism genes • NAT2, isoniazid anti-tuberculosis drug hepatotoxicity • CYP3A5, many drugs • Thiopurine S-methyltransferase (TPMT), 6-thioguanine • Drug targets (receptors) • B2 Adrenergic Receptor, inhaled B agonists for asthma • Drug transporters • P-glycoprotein (ABCB1, MDR1), resistance to antiepileptic drugs • The examples known today are those that come closest to simple genetic traits Potential Consequences • • • • • • Extended / shortened pharmacological effect Adverse drug reactions Lack of pro-drug activation Increased / decreased effective dose Metabolism by alternative, deleterious pathways Exacerbated drug-drug interactions The Goal of Pharmacogenomics Picture from Perlegen website: www.perlegen.com Complex Genetics: Concepts • • • • • • • Family studies vs. population studies Penetrance Genetic heterogeneity Linkage vs. association Haplotypes in family and association studies Genetic variation, SNPs Genotyping Types of Genetic Studies • Family studies – multi-generation families • Association studies – Case / control (easiest to collect) Penetrance • Penetrance = the proportion of carriers who show the phenotype • Expressivity = severity of the phenotype Genetic Heterogeneity • Locus heterogeneity (what we usually refer to when we talk about genetic heterogeneity) • Allelic heterogeneity Family Studies Identify Highly Penetrant Mutations High penetrance disease allele(s) Availability of suitable families is the limiting factor Family studies are effective for only a minority of conditions Association Studies Can Identify Variants with High or Low Penetrance • Case / control groups • Not limited to high penetrance alleles • Amenable to the study of gene-environment interactions • A preferred approach for the majority of complex genetic disorders Complex Diseases / Phenotypes • • • • Multigenic (genetic heterogeneity) Environmental effects (multiple) Gene-gene interactions Gene-environment interactions (for pharmacogenetic traits: age, alcohol consumption, hepatitis exposure, etc.) • Association studies will hold up under these complications but family-based linkage studies will not! Linkage vs. Association • Linkage is to a locus – different families can be linked to the same locus but have different disease alleles – how to take advantage of this in proving a gene is responsible for a disease • Association is with an allele – done in groups or populations – the allele arose and was propagated in the population; the haplotype was degraded by recombination Genetic Markers SNPs: Substitutions, for example, C / T Most common type of genetic variation Ideal for association mapping over short distances 1 SNP every ~ 200 base pairs in a population 1 SNP every ~1000 base pairs between 2 individuals dbSNP: >10M putative SNPs, > 5M validated SNPs Microsatellites: (CA)n or other short repeats More polymorphic than SNPs Less common than SNPs 1 polymorphic microsatellite per ~ 100,000 base pairs Best for linkage mapping over long distances, in families SNPs • Single Nucleotide Polymorphisms • Can also use “Indels”, though some investigators throw them away! • Synonymous, non-synonymous SNPs • Mutation vs. polymorphism vs. variant or variation • The 1% definition SNP Databases • • • • • • dbSNP (more than just human) Human Genome Variation Database At least 11 others! ~ 10 million SNPs with minor allele >1% ~ 7 million SNPs with minor allele >5% ~ 50,000 non-synonymous SNPs in the human genome Case / Control Studies 1. 2. 3. 4. 5. 6. 7. Collect blood samples from patients and controls, with consent Establish database of clinical and epidemiological data Select ‘candidate’ genes of interest for each trait Sequence the candidate genes in a small group of patients Genotype selected variants in case / control groups Analyze for association with a phenotype Analyze for gene-gene and gene-environment interactions Genetic, Ethical, Legal and Social (GELS) issues investigations Linkage Disequilibrium • The difference between the observed frequency of a haplotype and its expected frequency if all alleles were segregating randomly • For adjacent loci: A,a B,b • D = PAB - PA x PB • D is dependent on allele frequencies • Other related measures also used Human haplotype blocks . . . Ancestral chromosomes Observed pattern of historical recombination in common haplotypes Rather than 50 kb . . . Simplify association studies Ancestral chromosomes A disease-causing mutation arises Association with nearby SNPs SNP1 SNP2 A C A C G T G T A CA * A C G TG G T G CA * A T A TG G C Location of mutation Gene LD and Association • Direct association – asks about the effect of a variant – if negative, the gene may still be involved! • Indirect association – uses LD – can be more convincingly negative if haplotypes are assessed Haplotype Blocks • • • • Became clear in October 2001 87% of the genome is in blocks ~> 30 kb Not all of the genome is in haplotype blocks! Average block 22 kb, 11kb in African populations (Gabriel et al, 2002) • A few common haplotypes at a given locus in a given population • African populations generally have the greatest number of haplotypes and the shortest haplotype blocks • Strength of LD and size of blocks varies greatly between regions How to Generate Haplotypes • Haplotyping in families • Physical determination – long-range PCR, separation of molecules – cloning of single molecules – labor intensive • Estimate haplotype frequencies – Expectation Maximization algorithm, others – generate frequencies for case group, control group Tag SNPs Chromosome copy 1 Chromosome copy 2 Chromosome copy 3 Chromosome copy 4 The HapMap • Reference map for association studies • Expected to reduce the number of markers required to conduct effective genome scans for association • 270 samples from 4 populations: – – – – 30 Yoruban trios (Nigeria) 45 unrelated Japanese (Tokyo) 45 unrelated Chinese (Beijing) 30 U.S. trios (CEPH, N/W European ancestry) • >400,000 markers genotyped in all samples, nearly 1M in CEPH trios Strategies • Candidate gene based studies – hypothesis-driven – must guess (one of) the right gene(s)!! – Current state of the art • Genome scans – “hypothesis-free” – scans of ~ 1 million markers are now possible SNP Discovery is Still Necessary • Many have been found by multi-read sequence mining • Directed public SNP discovery in certain sets of genes, e.g.: – SNP500Cancer – Environmental Genome Project (EGP) • Individuals used usually “unaffected” SNP Discovery All exons and regulatory regions of each gene Identify regulatory regions by comparative genomics Bi-directional sequencing Denaturing High Performance Liquid Chromatography (DHPLC) Other methods 1 2 3 PCR Set-up: Packard Multiprobe II liquid handler Template aliquotting: Robbins Hydra PCR and cycle sequencing: MJ Tetrads 5 4 6 Sequencing: ABI 3700s Purification of PCR Products: Agencourt SNP Discovery: PolyPhred and Consed PolyPhred: Debbie Nickerson; Consed, Phil Green Sample Output GG GA AA Genotyping, Technology • Determining the allele(s) present in a particular sample at a particular (SNP) marker • Many methods TaqMan (ABI): Uniplex genotyping TaqMan TaqMan Output Homozygous 1,1 Heterozygous Homozygous 2,2 MassEXTEND REACTION Allele 1 Allele 2 Unlabeled Primer (23-mer) Same Primer (23-mer) TCT ACT +Enzyme +ddATP +dCTP/dGTP/dTTP Extended Primer (26-mer) Diagram courtesy of Sequenom Allele 2 Allele 2 Allele 1 EXTEND Primer TG A ACT A TCT EXTEND Primer Allele 1 EXTEND Primer Extended Primer (24mer) Sequenom MassARRAY: < 12-plex * T C * * A C T G * A G * A G Diagram courtesy of Sequenom Illumina BeadArray System: 1152-plex • 1152-fold multiplexing • 0.26 ng of genomic DNA per genotype • $ 0.05 USD per genotype Total Internal Reflection Fiber Cladding Photons (out) Fiber Core Photons (in) Fluorescence Emission Excitation Beam cladding Illumina BeadArray System B A Decoder Oligo Decode hyb 1 Decode hyb 2 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 T/C P1’ P2’ P1 P2 A G Address’ P3’ PCR with common primers P3 /\/\/\/ Address Decode hyb. 1 Decode hyb. 2 Allele Specific Extension Product capture by hybridization to array ParAllele Molecular Inversion Probes: 10,000 Plex Affymetrix Whole Genome Sampling Analysis: 500,000-plex Kennedy et al., 2003 Affymetrix: Allele-Specific Hybridization PM = perfect match MM = mismatch DNA Pooling Strategies • Reduce the number of genotypes and genotyping cost, particularly for whole genome scans • Pool of case DNAs vs. pool of control DNAs • DNAs must be mixed in precisely equimolar proportions in the pools! • Requires a quantitative genotyping technique • E.g. 40% in cases vs. 20% in controls • Verify positives by genotyping individual samples