* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Linkage group on OL
Comparative genomic hybridization wikipedia , lookup
Primary transcript wikipedia , lookup
Public health genomics wikipedia , lookup
DNA damage theory of aging wikipedia , lookup
Cancer epigenetics wikipedia , lookup
DNA barcoding wikipedia , lookup
Metagenomics wikipedia , lookup
United Kingdom National DNA Database wikipedia , lookup
DNA vaccination wikipedia , lookup
Molecular Inversion Probe wikipedia , lookup
DNA profiling wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Genetic engineering wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Point mutation wikipedia , lookup
Nucleic acid double helix wikipedia , lookup
SNP genotyping wikipedia , lookup
Gel electrophoresis of nucleic acids wikipedia , lookup
DNA supercoil wikipedia , lookup
Extrachromosomal DNA wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Non-coding DNA wikipedia , lookup
Genetically modified crops wikipedia , lookup
Quantitative trait locus wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Epigenomics wikipedia , lookup
Cell-free fetal DNA wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Bisulfite sequencing wikipedia , lookup
Molecular cloning wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Designer baby wikipedia , lookup
Genomic library wikipedia , lookup
Genealogical DNA test wikipedia , lookup
Microevolution wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Genome editing wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Microsatellite wikipedia , lookup
History of genetic engineering wikipedia , lookup
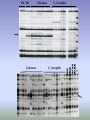
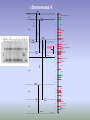
ISSAG Viterbo - 22 / 26 August 2005 CONTRIBUTION TO FINE MAPPING OF OL-2 LOCUS IN TOMATO MINOIA SILVIA Department of Agro-forestry and Environmental Biology and Chemistry Sect. Genetics and Plant Breeding, University of Bari (Italy) Powdery mildew caused by Oidium lycopersici on tomato’s leaves L. esculentum cv.SuperMarmande: SM Susceptible L. esculentum var. cerasiforme: R28 Resistant Susceptible Resistant Sources of resistance to the fungus are available in wild material and in particular in the accession of L. esculentum var. cerasiforme,we have selected a line (LC-95) resulting in complete resistance to powdery mildew, and also showed that a single recessive gene, named ol-2, was responsible for desease control. Our first step was isolated a RAPD marker (OPU31500) linked to ol-2 gene to using a “Bulked Segregant Analysis” (BSA): it was detected in the susceptible bulk. The OPU31500 was converted to a CAPS marker and the estimation of the distance between the marker and the ol-2 gene was identified by linkage analysis in the F2 population. SM R28 PS PR F2 Resistant F2 Susceptible Piante F2 bulk suscettibile M 270bp F2 Resistant F2 Susceptible F1 Electrophoretic patterns of PCR-amplified DNA products obtained with OPU3 primer 5'-CTATGCCGA-3' from genomic DNA of parents (SM susceptible; R28 resistant), susceptible F1 plants, bulks (BLKS, bulk of F2 susceptible plants; BLKR, bulk of F2 resistant plants) and all the individuals included in the bulks (lanes 1-9 susceptible; lanes 10-19 resistant): the absence of the 1.5 kb band, indicated with the arrow and designated OPU31500, is associated with resistance. M1 and M2, DNA molecular weight markers (1kb and 100bp Ladders, respectively) chromosome 4 IL4-1-1 +GP180 4-A GP180 3.7 4-B -TG15 +CT229A IL4-1 TG15,CT229A 12.1 IL4-2 4.6 (CT269B,TG123) TG483 3.4 Tpi-2 4.3 -CT175 +TG182 1.1 0.8 +TG182 -CT157 IL4-3-2 5.5 4-E 4-F (CD49A) TG370 4-C 4-D (CT122C,CT63C,TG413B,TG49, 6Pgdh-1) CD59 2.9 3.9 2.4 +TG208 -TG75A TG474,TG339 CT175 TG182,CT192 (CD70,TG146) CT157 (TG609,CT162,PC1, Pgm-2, Got-1) TG506,TG2 (GP221,TG316,CT261,TG268,CT181,CT145B) TG652,CT259 3.6 (CT97,TG516) 1.2 2.1 0.8 TG287 (TG633,PC3,CT178) TG208 (TG272B,TG95, Adh-1) TG75A CD55 +CD55 -TG519 6.0 (CT286) TG519,TG264,TG635,CT194 4.6 (CT185) IL4-3 TG62,TG427,CT161 8.9 (CT188) TG65 4-G 8.0 (TG120) 3.5 TG574 (TG305,CT132A, CT264) TG555 2.1 TG155 9.4 -TG155 +CT50 CT287B 5.6 1.0 1.0 4-H 3.5 (TG34) CT50 TG500 CT133 (TG443) CD39,CT73 IL4-4 4.9 +CP57 -CT173 2.3 (TG260) CP57 1.6 CT173 TG22,CT126B 5.8 (CT253,CT239A,TG37X) 4-I TG163,CT224B,CT199 3.8 (CT61) TG464,TG498,TG587 +TG464 Our second step was to identify AFLP markers always associated at the ol-2 gene. We found 8 new molecular markers. E39/M37(290) Linkage group on Ol-2 locus 3.2 E32/M47(174) E32/M50(174) 0.4 E40/M56(100) E34/M61(270) E42/M45(248) 0.2 E41/M32(390) 0.3 0.5 OPU3 ol-2 CAPS/OPU3 cM E32/M53(280) The finale purpose of our investigation is to identify molecular markers linked to resistance and to use these markers for MAS (marker assistent selection), to set up improved lines of tomato resistant to powdery mildew (Oidium lycopersicum). RESEARCH AIMS To identify new PCR-based molecular markers linked to ol-2 gene To establish a linkage group for Ol-2 locus To obtain co-dominant markers useful to perform MAS in tomato: in particular to converted the 8 AFLP marker in SCAR or CAPS markers. 3 2 1 EcoRI 41/MseI 32 EcoRI 34/MseI 61 4 3 2 1 4 3 2 DNA Marker EcoRI 32/MseI 53 Lambda DNA ng/µl 90 60 30 1 EcoRI 34/MseI 61 (2) 4 3 2 1 4 SCAR MARKERS SCAR markers obtained from conversion of new AFLP polymorphic markers Legend: SM= 1; R28=2; BLKS=3; BLKR=4 DNA Marker= 25bp When we begins work with this CAPS markers started our problems: • the fragments that we amplified were small (between 100-300 bp) • when we cutted with restiction enzymes we obteined smaller fragments and we lost the polymorphism. LMS-PCR technique (Schupp et al., 1999) (Ligation-Mediated Suppression PCR) this is an systematic approach to obtain, from AFLP markers, information about internal and flanking sequence. ‘Wolking’ on the genome, we arrive to amplify unknown regions flanking known AFLP regions and then to lengthen our fragments. Starting from 100-300 bp fragments we obteined fragments of 1000 bp. Our work is in progress… … THANK’S FOR YOUR ATTENTION Michelmore RW, Paran RV, Kesseli, Identification of marker linked to desease resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating population, Proc. Natl. Acad. Sci. USA 88 (1991) 9828-9833. Schupp JM, Price LB, Klevytska A and Keim P, 1999. Internal and flanking sequence from AFLP fragment using ligation-mediated suppression PCR. BioTechniques 26, 905-912. Steps of LMS-PCR technique: • to digest genomic DNA • to attach adaptors • to amplify it using primers of known sequence and adaptor primers